Predicate Oriented Pattern Analysis for Biomedical Knowledge Discovery.
引用次数: 16
Abstract
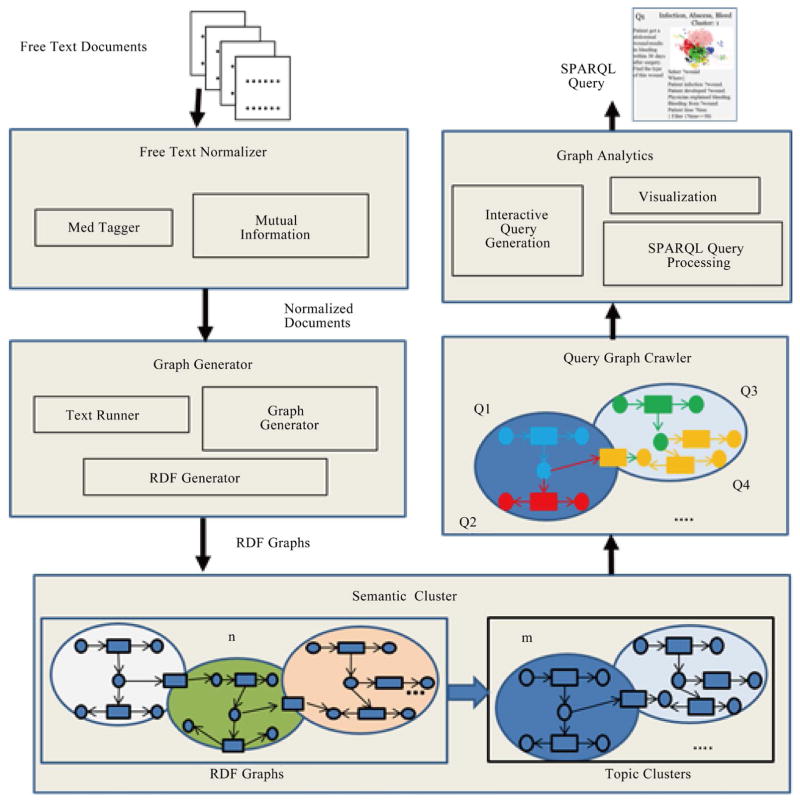
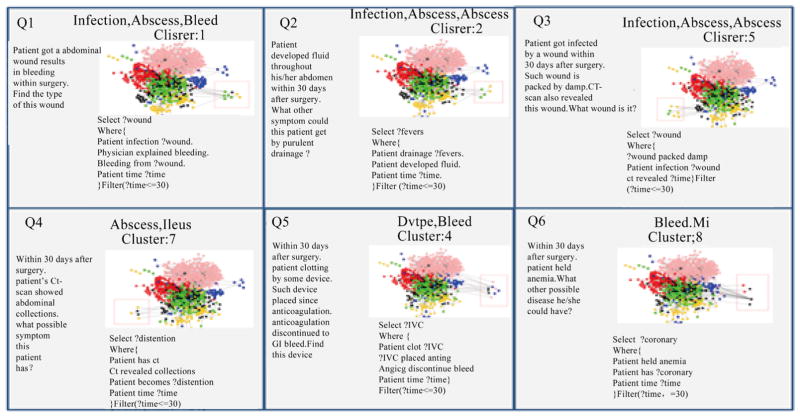
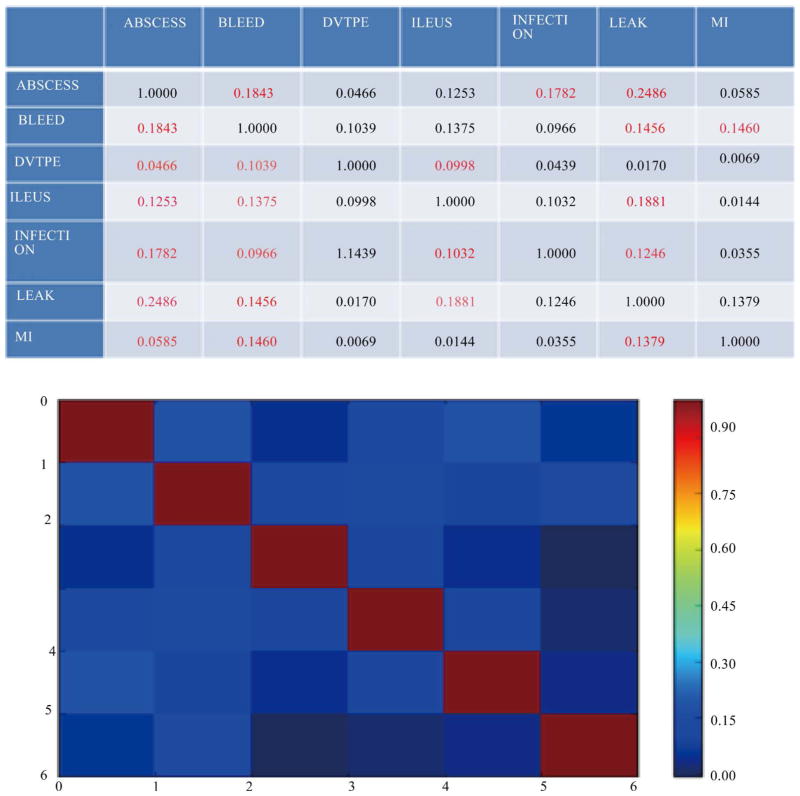
In the current biomedical data movement, numerous efforts have been made to convert and normalize a large number of traditional structured and unstructured data (e.g., EHRs, reports) to semi-structured data (e.g., RDF, OWL). With the increasing number of semi-structured data coming into the biomedical community, data integration and knowledge discovery from heterogeneous domains become important research problem. In the application level, detection of related concepts among medical ontologies is an important goal of life science research. It is more crucial to figure out how different concepts are related within a single ontology or across multiple ontologies by analysing predicates in different knowledge bases. However, the world today is one of information explosion, and it is extremely difficult for biomedical researchers to find existing or potential predicates to perform linking among cross domain concepts without any support from schema pattern analysis. Therefore, there is a need for a mechanism to do predicate oriented pattern analysis to partition heterogeneous ontologies into closer small topics and do query generation to discover cross domain knowledge from each topic. In this paper, we present such a model that predicates oriented pattern analysis based on their close relationship and generates a similarity matrix. Based on this similarity matrix, we apply an innovated unsupervised learning algorithm to partition large data sets into smaller and closer topics and generate meaningful queries to fully discover knowledge over a set of interlinked data sources. We have implemented a prototype system named BmQGen and evaluate the proposed model with colorectal surgical cohort from the Mayo Clinic.
面向谓词的生物医学知识发现模式分析。
在当前的生物医学数据运动中,已经进行了大量的努力,将大量传统的结构化和非结构化数据(例如,电子病历、报告)转换和规范化为半结构化数据(例如,RDF、OWL)。随着越来越多的半结构化数据进入生物医学领域,异构领域的数据集成和知识发现成为重要的研究问题。在应用层面,医学本体之间相关概念的检测是生命科学研究的重要目标。更重要的是,通过分析不同知识库中的谓词,弄清楚不同概念如何在单个本体内或跨多个本体关联。然而,当今世界是一个信息爆炸的世界,如果没有模式分析的支持,生物医学研究人员很难找到现有的或潜在的谓词来实现跨领域概念之间的链接。因此,需要一种机制来进行面向谓词的模式分析,以将异构本体划分为更紧密的小主题,并进行查询生成以从每个主题中发现跨领域知识。在本文中,我们提出了这样一个模型,基于它们的密切关系来预测面向模式分析,并生成相似矩阵。基于该相似矩阵,我们应用一种创新的无监督学习算法将大数据集划分为更小、更紧密的主题,并生成有意义的查询,以在一组相互关联的数据源上充分发现知识。我们已经实现了一个名为BmQGen的原型系统,并与梅奥诊所的结直肠手术队列一起评估了所提出的模型。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


