Diversity of Multidrug Efflux Genes and Phenotypic Evaluation of the In vitro Resistance Dynamics of Clinical Staphylococcus Aureus Isolates Using Methicillin; a Model β-lactam.
Q3 Immunology and Microbiology
Open Microbiology Journal
Pub Date : 2017-06-30
eCollection Date: 2017-01-01
DOI:10.2174/1874285801711010132
引用次数: 12
Abstract
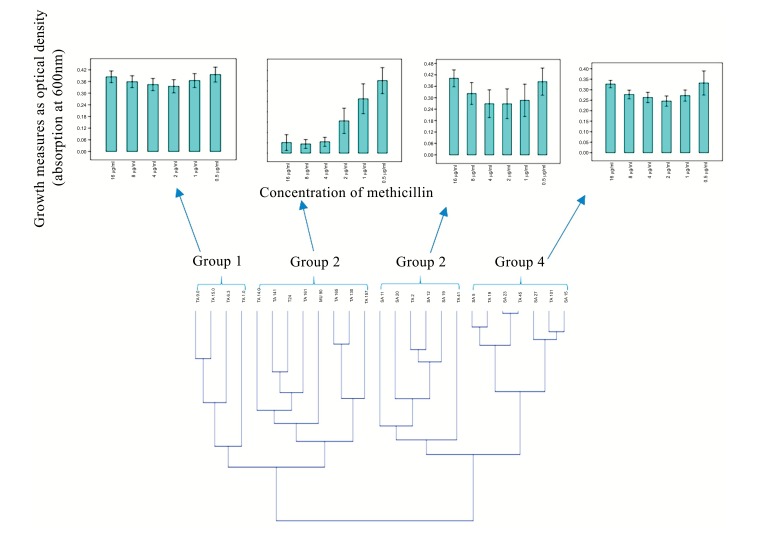
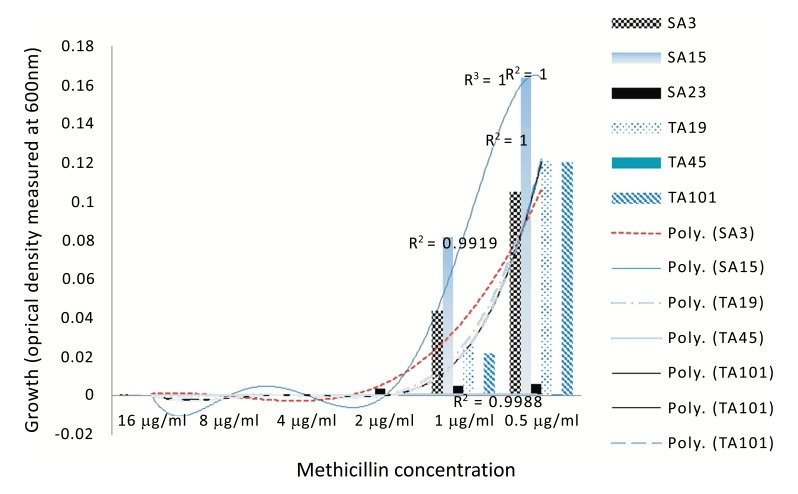
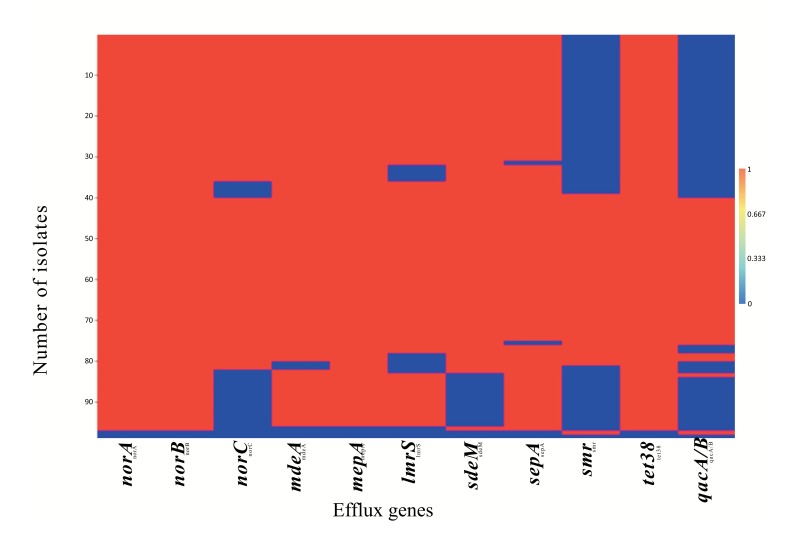
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) across the world often leave clinicians with little or no choice of treatment options. The multi-drug efflux (MDE) genes are bacterial survival mechanisms responsible for the pumping out of antibiotics and other biocides from the cytoplasm. Whilst effort is being made in the development of antibiotic adjuvants such as efflux pumps inhibitors, information is needed on the diversity of these MDEs in the circulating S. aureus and on the growth dynamics of the clinical isolates in response to antibiotics is not regularly examined. Methods: Here, we evaluated the diversity of MDEs in cinical S. aureus recovered in a tertiary academic hospital, Pretoria, South African hospital using PCR and also employed visual minimum inhibitory concentration and quantitative analysis of spectrophometric measurements of bacterial growth in the presence of a model β lactam antibiotic (methicillin), to phenotypically elucidate the resistance pattern of these isolates in response to methicillin. Results: Three major distribution patterns of MDEs were observed in the clinical isolates evaluated. Moreover, norA, nor B and tet38 were present in 98.9% of the isolates while other MDE were present in different proportions ranging from 40 to 98.6% of the isolates. In addition, S. aureus isolates, be it of MRSA or MSSA genotype did not habour the same set of MDEs despite being recovered from the same hospital setting. Finally, we showed that MSSA displayed phenotypic resistance to methicilllin despite the non-detection of the mecA resistance gene. Conclusions: Our data suggest that the growth of S. aureus may be enhanced by β lactams (methicillin) and that MSSA may also display resistance to methicillin and perhaps other β lactam antibiotics. The high prevalence of MDEs suggestive of resistance to a broad spectrum of biocides and fluoroquinolones are particularly disturbing.
多药外排基因多样性及临床分离金黄色葡萄球菌甲氧西林体外耐药动力学的表型评价a模型β-内酰胺。
目的:耐甲氧西林金黄色葡萄球菌(MRSA)在世界各地往往留给临床医生很少或根本没有选择的治疗方案。多药外排(MDE)基因是细菌的生存机制,负责从细胞质中泵出抗生素和其他杀菌剂。虽然正在努力开发抗生素佐剂,如外排泵抑制剂,但需要了解循环中的金黄色葡萄球菌中这些MDEs的多样性,以及临床分离株对抗生素反应的生长动态,这些信息没有定期检查。方法:本研究采用PCR方法对南非比勒陀利亚某三级学术医院临床金黄色葡萄球菌MDEs的多样性进行了评估,并采用视觉最小抑制浓度和细菌生长的分光光度测定法对模型β内酰胺抗生素(甲氧西林)进行了定量分析,以表型方式阐明这些菌株对甲氧西林的耐药模式。结果:临床分离株中MDEs主要有三种分布模式。此外,98.9%的分离株中存在norA、nor B和tet38,而其他MDE的存在比例从40%到98.6%不等。此外,金黄色葡萄球菌分离株,无论是MRSA还是MSSA基因型,尽管从同一医院环境中恢复,但并不具有相同的MDEs组。最后,我们发现尽管没有检测到mecA抗性基因,但MSSA对甲氧西林表现出表型抗性。结论:我们的数据表明,金黄色葡萄球菌可能被β内酰胺(甲氧西林)促进生长,并且MSSA也可能对甲氧西林和其他β内酰胺类抗生素产生耐药性。MDEs的高流行率表明对广泛的杀菌剂和氟喹诺酮类药物具有耐药性,这尤其令人不安。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Open Microbiology Journal
Immunology and Microbiology-Immunology and Microbiology (all)
CiteScore
1.80
自引率
0.00%
发文量
24
期刊介绍:
The Open Microbiology Journal is a peer-reviewed open access journal which publishes research articles, reviews/mini-reviews, case studies, guest edited thematic issues and short communications/letters covering theoretical and practical aspects of Microbial systematics, evolutionary microbiology, immunology, virology, parasitology , bacteriology, mycology, phycology, protozoology, microbial ecology, molecular biology, microbial physiology, biochemistry, microbial pathogenesis, host-microbe interaction, systems microbiology, synthetic microbiology, bioinformatics. The Open Microbiology Journal , a peer-reviewed journal, is an important and reliable source of current information on developments in the field. The emphasis will be on publishing quality papers rapidly and freely available to researchers worldwide.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


