Genome sequence of the white-rot fungus Irpex lacteus F17, a type strain of lignin degrader fungus.
Q3 Biochemistry, Genetics and Molecular Biology
Standards in Genomic Sciences
Pub Date : 2017-09-12
eCollection Date: 2017-01-01
DOI:10.1186/s40793-017-0267-x
引用次数: 16
Abstract
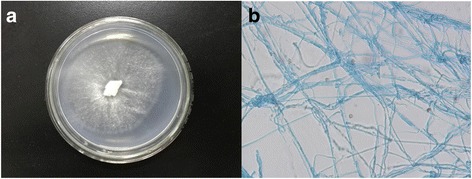
Irpex lacteus, a cosmopolitan white-rot fungus, degrades lignin and lignin-derived aromatic compounds. In this study, we report the high-quality draft genome sequence of I. lacteus F17, isolated from a decaying hardwood tree in the vicinity of Hefei, China. The genome is 44,362,654 bp, with a GC content of 49.64% and a total of 10,391 predicted protein-coding genes. In addition, a total of 18 snRNA, 842 tRNA, 15 rRNA operons and 11,710 repetitive sequences were also identified. The genomic data provides insights into the mechanisms of the efficient lignin decomposition of this strain.

木质素降解真菌型菌株白腐菌Irpex lacteus F17的基因组序列。
Irpex lacteus是一种世界性的白腐菌,降解木质素和木质素衍生的芳香化合物。在这项研究中,我们报道了从中国合肥附近的一棵腐烂的硬木树上分离到的高质量的I. lacteus F17基因组序列草图。基因组长度为44,362,654 bp, GC含量为49.64%,共预测蛋白编码基因10,391个。此外,还鉴定出18个snRNA、842个tRNA、15个rRNA操纵子和11710个重复序列。基因组数据提供了深入了解该菌株的有效木质素分解机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Standards in Genomic Sciences
GENETICS & HEREDITY-MICROBIOLOGY
CiteScore
1.44
自引率
0.00%
发文量
0
审稿时长
6-12 weeks
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


