Nucleotide Interdependency in Transcription Factor Binding Sites in the Drosophila Genome.
Gene regulation and systems biology
Pub Date : 2016-06-12
eCollection Date: 2016-01-01
DOI:10.4137/GRSB.S38462
引用次数: 3
Abstract
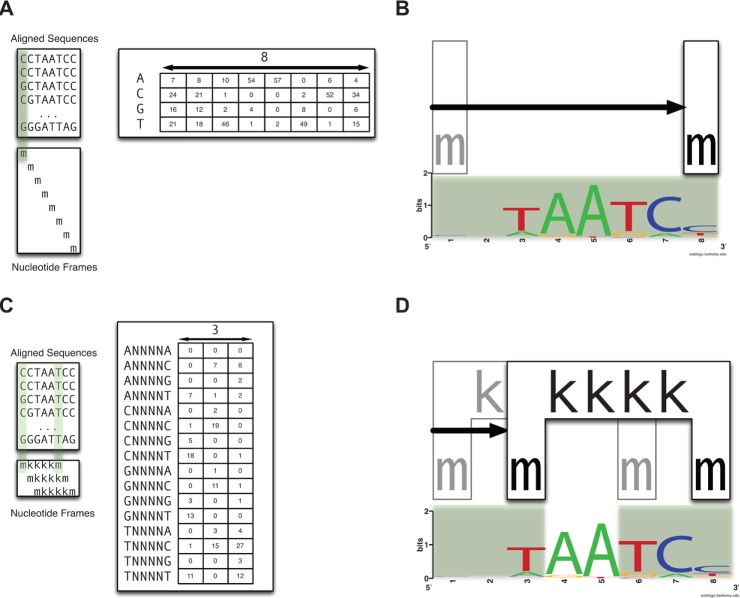
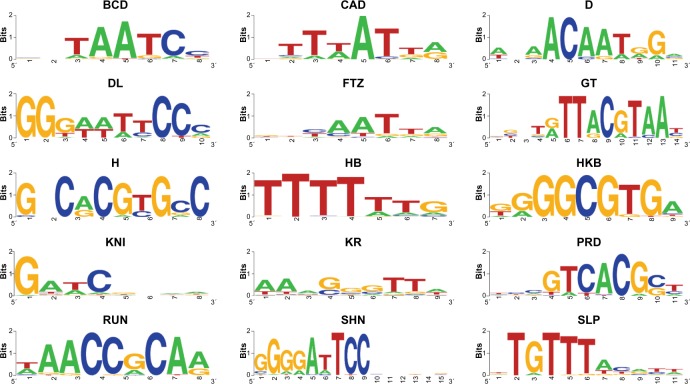
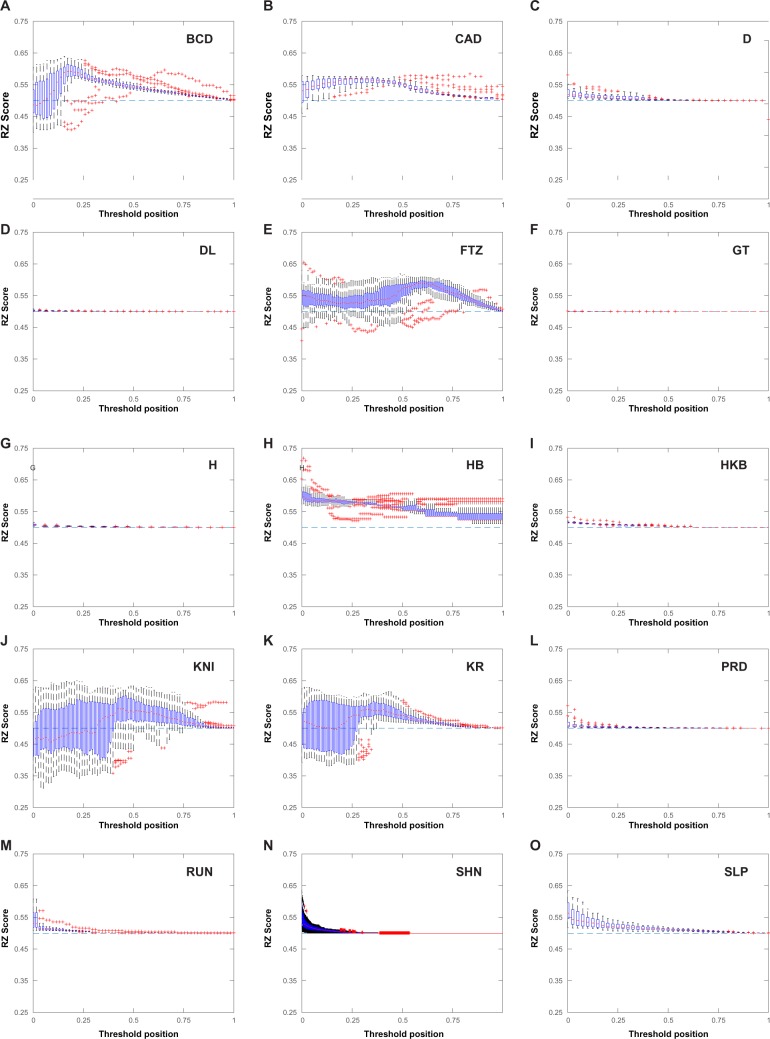
A long-standing objective in modern biology is to characterize the molecular components that drive the development of an organism. At the heart of eukaryotic development lies gene regulation. On the molecular level, much of the research in this field has focused on the binding of transcription factors (TFs) to regulatory regions in the genome known as cis-regulatory modules (CRMs). However, relatively little is known about the sequence-specific binding preferences of many TFs, especially with respect to the possible interdependencies between the nucleotides that make up binding sites. A particular limitation of many existing algorithms that aim to predict binding site sequences is that they do not allow for dependencies between nonadjacent nucleotides. In this study, we use a recently developed computational algorithm, MARZ, to compare binding site sequences using 32 distinct models in a systematic and unbiased approach to explore nucleotide dependencies within binding sites for 15 distinct TFs known to be critical to Drosophila development. Our results indicate that many of these proteins have varying levels of nucleotide interdependencies within their DNA recognition sequences, and that, in some cases, models that account for these dependencies greatly outperform traditional models that are used to predict binding sites. We also directly compare the ability of different models to identify the known KRUPPEL TF binding sites in CRMs and demonstrate that a more complex model that accounts for nucleotide interdependencies performs better when compared with simple models. This ability to identify TFs with critical nucleotide interdependencies in their binding sites will lead to a deeper understanding of how these molecular characteristics contribute to the architecture of CRMs and the precise regulation of transcription during organismal development.
果蝇基因组中转录因子结合位点的核苷酸相互依赖性。
现代生物学的一个长期目标是描述驱动生物体发育的分子成分。真核生物发育的核心是基因调控。在分子水平上,该领域的大部分研究都集中在转录因子(tf)与基因组中被称为顺式调控模块(CRMs)的调控区域的结合上。然而,对许多tf的序列特异性结合偏好知之甚少,特别是关于构成结合位点的核苷酸之间可能的相互依赖性。许多旨在预测结合位点序列的现有算法的一个特殊限制是,它们不允许非相邻核苷酸之间的依赖关系。在这项研究中,我们使用最近开发的计算算法MARZ,以系统和无偏倚的方法比较32种不同模型的结合位点序列,以探索已知对果蝇发育至关重要的15种不同tf的结合位点内的核苷酸依赖性。我们的研究结果表明,许多这些蛋白质在其DNA识别序列中具有不同水平的核苷酸相互依赖性,并且,在某些情况下,考虑这些依赖性的模型大大优于用于预测结合位点的传统模型。我们还直接比较了不同模型识别crm中已知KRUPPEL TF结合位点的能力,并证明了与简单模型相比,考虑核苷酸相互依赖性的更复杂模型表现更好。这种鉴定在其结合位点上具有关键核苷酸相互依赖性的tf的能力将使我们更深入地了解这些分子特征如何促进crm的结构和在生物体发育过程中精确调节转录。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


