Identification of cis-Regulatory Elements in the dmyc Gene of Drosophila Melanogaster.
Gene regulation and systems biology
Pub Date : 2012-01-01
Epub Date: 2011-12-21
DOI:10.4137/GRSB.S8044
引用次数: 2
Abstract
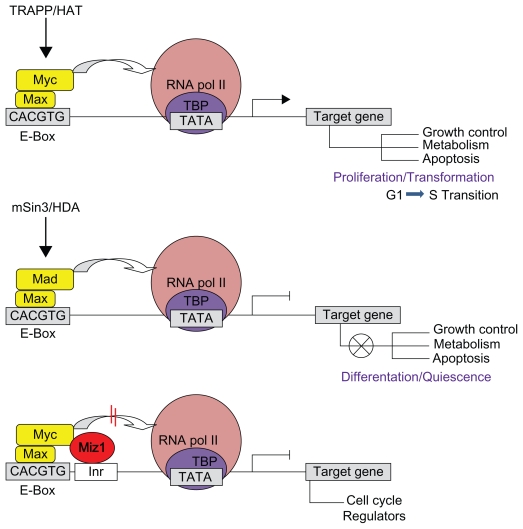
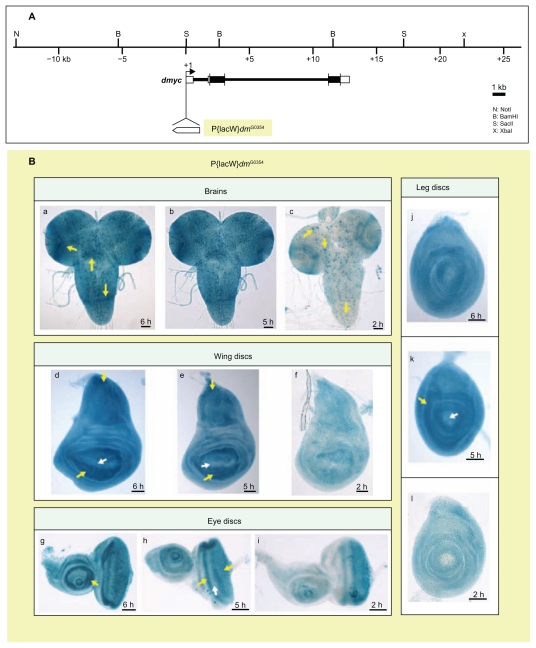
Myc is a crucial regulator of growth and proliferation during animal development. Many signals and transcription factors lead to changes in the expression levels of Drosophila myc, yet no clear model exists to explain the complexity of its regulation at the level of transcription. In this study we used Drosophila genetic tools to track the dmyc cis-regulatory elements. Bioinformatics analyses identified conserved sequence blocks in the noncoding regions of the dmyc gene. Investigation of lacZ reporter activity driven by upstream, downstream, and intronic sequences of the dmyc gene in embryonic, larval imaginal discs, larval brain, and adult ovaries, revealed that it is likely to be transcribed from multiple transcription initiation units including a far upstream regulatory region, a TATA box containing proximal complex and a TATA-less downstream promoter element in conjunction with an initiator within the intron 2 region. Our data provide evidence for a modular organization of dmyc regulatory sequences; these modules will most likely be required to generate the tissue-specific patterns of dmyc transcripts. The far upstream region is active in late embryogenesis, while activity of other cis elements is evident during embryogenesis, in specific larval imaginal tissues and during oogenesis. These data provide a framework for further investigation of the transcriptional regulatory mechanisms of dmyc.

果蝇dmyc基因顺式调控元件的鉴定。
Myc是动物发育过程中生长和增殖的关键调节因子。许多信号和转录因子导致果蝇myc表达水平的变化,但没有明确的模型来解释其在转录水平上调控的复杂性。在这项研究中,我们使用果蝇遗传工具来追踪dmyc顺式调控元件。生物信息学分析确定了dmyc基因非编码区域的保守序列块。对胚胎、幼虫影像盘、幼虫脑和成虫卵巢中由dmyc基因的上游、下游和内含子序列驱动的lacZ报告基因活性的研究表明,它可能是从多个转录起始单元转录的,包括远上游调控区、包含近端复合物的TATA盒和与内含子2区域内的启动子结合的TATA-less下游启动子元件。我们的数据为dmyc调控序列的模块化组织提供了证据;这些模块很可能需要生成dmyc转录本的组织特异性模式。远上游区域在胚胎发生晚期活跃,而其他顺式元件在胚胎发生、特定的幼虫想象组织和卵发生期间的活动是明显的。这些数据为进一步研究dmyc的转录调控机制提供了框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


