Shared Genomics: Developing an accessible integrated analysis platform for Genome-Wide Association Studies.
Summit on translational bioinformatics
Pub Date : 2010-03-01
引用次数: 0
Abstract
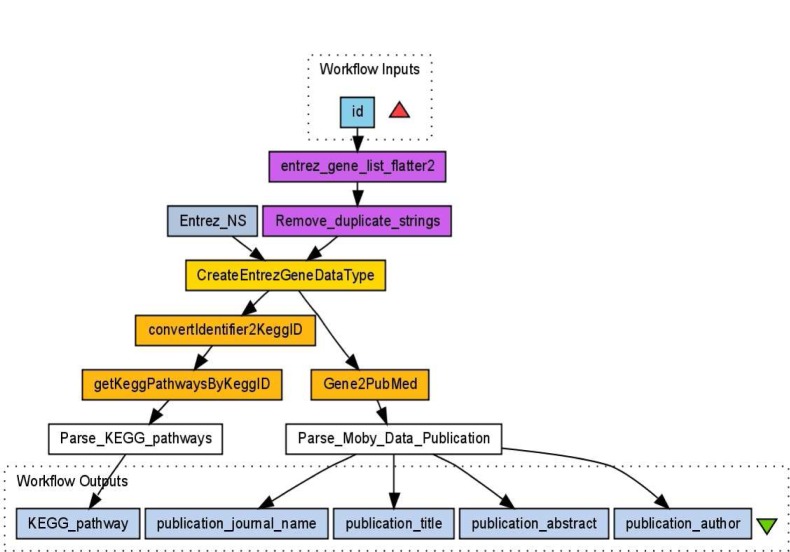
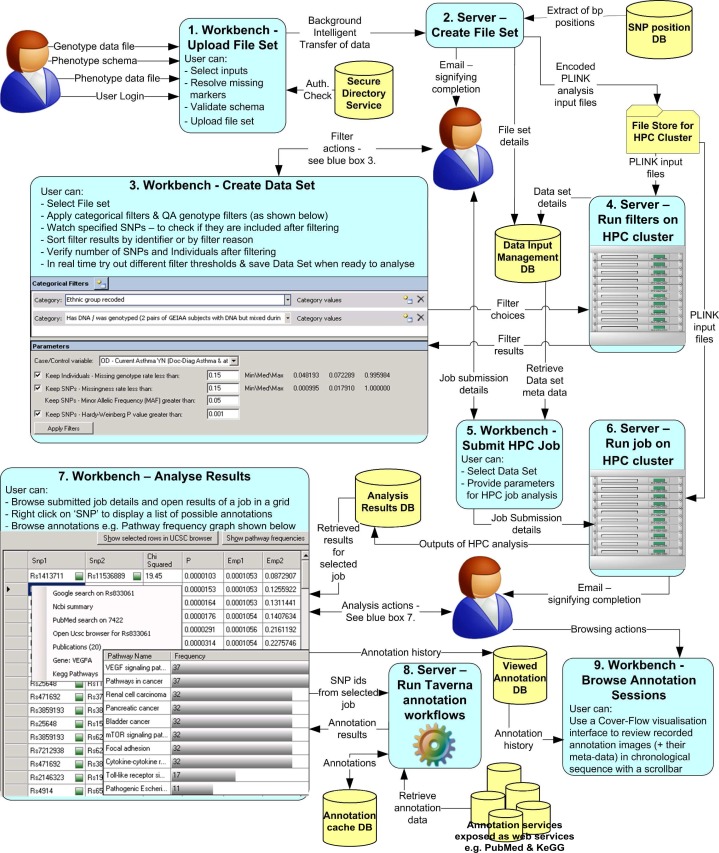
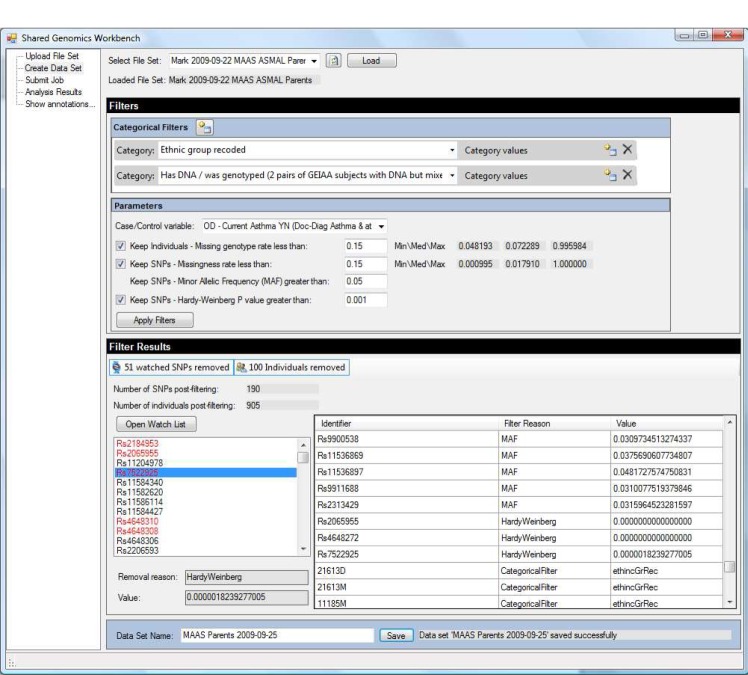
Increasingly, genome-wide association studies are being used to identify positions within the human genome that have a link with a disease condition. The number of genomic locations studied means that computationally intensive and bioinformatic intensive solutions will have to be used in the analysis of these data sets. In this paper we present an integrated Workbench that provides user-friendly access to parallelized statistical genetics analysis codes for clinical researchers. In addition we biologically annotate statistical analysis results through the reuse of existing bionformatic Taverna workflows.
共享基因组学:为全基因组关联研究开发一个可访问的集成分析平台。
越来越多的全基因组关联研究被用于确定人类基因组中与疾病状况有联系的位置。所研究的基因组位置的数量意味着必须使用计算密集型和生物信息学密集型的解决方案来分析这些数据集。在本文中,我们提出了一个集成的工作台,为临床研究人员提供用户友好的访问并行统计遗传学分析代码。此外,我们通过重用现有的生物构象Taverna工作流程对统计分析结果进行生物注释。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


