Multi-feature machine learning for enhanced drug–drug interaction prediction
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Drug–drug interactions are a major concern in healthcare, as concurrent drug use can cause severe adverse effects. Existing machine learning methods often neglect data imbalance and DDI directionality, limiting clinical reliability. To overcome these issues, we employed GPT-4o Large Language Model to convert free-text DDI descriptions into structured triplets for directionality analysis and applied SMOTE to alleviate class imbalance. Using four key drug features (molecular fingerprints, enzymes, pathways, targets), our Deep Neural Networks (DNN) achieved 88.9% accuracy and showed an average AUPR gain of 0.68 for minority classes attributable to SMOTE. By applying attention-based feature importance analysis, we demonstrated that the most influential feature in the DNN model was supported by pharmacological evidence. These results demonstrate the effectiveness of our framework for accurate and robust DDI prediction. The source code and data are available at https://github.com/FrankFengF/Drug-drug-interaction-prediction-
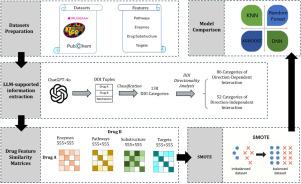
多特征机器学习增强药物-药物相互作用预测。
药物-药物相互作用是医疗保健中的一个主要问题,因为同时使用药物会导致严重的不良反应。现有的机器学习方法往往忽略了数据的不平衡和DDI的方向性,限制了临床的可靠性。为了克服这些问题,我们使用gpt - 40大型语言模型将自由文本DDI描述转换为结构化三元组进行方向性分析,并应用SMOTE来缓解类不平衡。利用四个关键的药物特征(分子指纹图谱、酶、途径、靶标),我们的深度神经网络(DNN)达到了88.9%的准确率,并且显示出归因于SMOTE的少数类别的平均AUPR增益为0.68。通过应用基于注意的特征重要性分析,我们证明DNN模型中最具影响力的特征得到了药理学证据的支持。这些结果证明了我们的框架对准确和稳健的DDI预测的有效性。源代码和数据可从https://github.com/FrankFengF/Drug-drug-interaction-prediction获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


