Taxonomic diversity of bacterial communities in the sediments of a soda lake in Arusha, Tanzania
IF 2.5
3区 环境科学与生态学
Q3 ENVIRONMENTAL SCIENCES
引用次数: 0
Abstract
Soda lakes are special ecosystems with high salinity and alkalinity, notably found in the Great Rift Valley of East Africa. Given the unique nature of soda lakes, it is interesting and valuable to examine their bacterial composition as an initial step towards bioprospecting. This study provides the first metagenomic snapshots of bacterial communities inhabiting Lake Natron sediments using the 16S rRNA gene sequenced using a PacBio sequencing system. Results show high abundance and diversity of species in general, with notable dominance of Firmicutes, Bacteroidota, Actinomycetota, and Proteobacteria with the relative abundances of 45.21 %, 25.23 %, 12.59 %, and 23.29 %, respectively. At the class level, Bacteroidia (23.54 %), Gammaproteobacteria (22.32 %), Bacilli (19.66 %), and Clostridia (22.32 %) were the most dominant classes. At lower taxonomic ranks (especially at the genus and species levels), there was an increase in the percentage of unknown and “Candidate Phyla” species (i.e., those discovered using genetic methods but not yet fully characterized or classified), suggesting the potential presence of new bacterial taxa in Lake Natron. The diversity indices revealed a high level of community diversity, with a large number of species, the presence of rare species, and an even distribution of bacteria across the sampling points. Although this study provides the first report on the existence of different bacterial taxa in Lake Natron, additional investigation into the biotechnological importance of the found species is of importance; hence, a functional metagenomic study is advised.
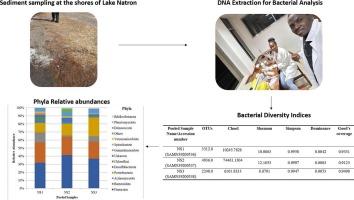
坦桑尼亚阿鲁沙苏打湖沉积物中细菌群落的分类多样性
苏打湖是一种特殊的高盐度和高碱度生态系统,主要分布在东非大裂谷。鉴于苏打湖的独特性质,研究其细菌组成作为生物勘探的第一步是有趣和有价值的。本研究利用PacBio测序系统对16S rRNA基因进行测序,提供了栖息在Natron湖沉积物中的细菌群落的第一个宏基因组快照。结果表明,总体上物种丰度和多样性较高,其中厚壁菌门、拟杆菌门、放线菌门和变形菌门优势显著,相对丰度分别为45.21%、25.23%、12.59%和23.29%。在纲水平上,Bacteroidia(23.54%)、Gammaproteobacteria(22.32%)、Bacilli(19.66%)和Clostridia(22.32%)为优势纲。在较低的分类等级(特别是属和种水平)上,未知和“候选门”物种(即使用遗传方法发现但尚未完全表征或分类的物种)的百分比有所增加,表明Natron湖可能存在新的细菌分类群。群落多样性指数显示群落多样性水平高,物种数量多,存在稀有物种,细菌在各采样点分布均匀。虽然本研究首次报道了纳特龙湖不同细菌分类群的存在,但对所发现物种的生物技术重要性的进一步研究具有重要意义;因此,建议进行功能宏基因组研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Great Lakes Research
生物-海洋与淡水生物学
CiteScore
5.10
自引率
13.60%
发文量
178
审稿时长
6 months
期刊介绍:
Published six times per year, the Journal of Great Lakes Research is multidisciplinary in its coverage, publishing manuscripts on a wide range of theoretical and applied topics in the natural science fields of biology, chemistry, physics, geology, as well as social sciences of the large lakes of the world and their watersheds. Large lakes generally are considered as those lakes which have a mean surface area of >500 km2 (see Herdendorf, C.E. 1982. Large lakes of the world. J. Great Lakes Res. 8:379-412, for examples), although smaller lakes may be considered, especially if they are very deep. We also welcome contributions on saline lakes and research on estuarine waters where the results have application to large lakes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


