The cAMP receptor protein, SyCRP1 acts as a transcriptional repressor of CO2-concentrating mechanism genes at high inorganic carbon levels in Synechocystis PCC 6803
IF 3.1
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochimica et Biophysica Acta-Gene Regulatory Mechanisms
Pub Date : 2025-10-02
DOI:10.1016/j.bbagrm.2025.195117
引用次数: 0
Abstract
Cyanobacteria utilize a CO2-concentrating mechanism (CCM) to enhance photosynthetic efficiency by accumulating CO2 around RuBisCO, a process crucial for adapting to fluctuating environmental CO2 levels. While the upregulation of CCM genes under low inorganic carbon (Ci) conditions is known, the precise Ci sensing and regulatory mechanisms governing CCM gene expression remain incompletely understood. We show that a membrane-bound SyCRP1 senses high Ci levels through cAMP binding to its low- and high-affinity sites. We demonstrate its interaction with membrane lipids and liposomes, and a subsequent reversal of its membrane localization upon cAMP binding. Comprehensive ChIP-seq analysis reveals direct binding of SyCRP1 to regulatory elements of core CCM genes. Our findings establish SyCRP1 as a key transcriptional repressor of these genes under high Ci conditions, significantly advancing our understanding of the molecular mechanisms governing CCM genes' expression in cyanobacteria.
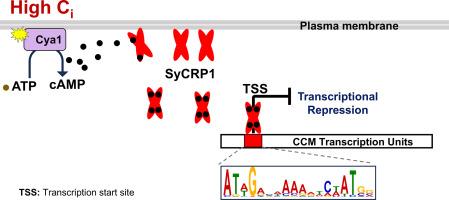
聚囊藻(Synechocystis PCC 6803)中,cAMP受体蛋白SyCRP1作为高无机碳水平下co2富集机制基因的转录抑制因子。
蓝藻利用二氧化碳浓缩机制(CCM)通过在RuBisCO周围积累二氧化碳来提高光合效率,这一过程对于适应环境中波动的二氧化碳水平至关重要。虽然已知CCM基因在低无机碳(Ci)条件下的上调,但CCM基因表达的精确Ci感知和调控机制仍不完全清楚。我们发现,膜结合的SyCRP1通过cAMP结合到其低和高亲和力位点来感知高Ci水平。我们证明了它与膜脂和脂质体的相互作用,以及随后在cAMP结合时其膜定位的逆转。综合ChIP-seq分析显示SyCRP1与核心CCM基因的调控元件直接结合。我们的研究结果表明,SyCRP1在高Ci条件下是这些基因的关键转录抑制因子,这极大地促进了我们对蓝藻中CCM基因表达的分子机制的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.20
自引率
2.10%
发文量
63
审稿时长
44 days
期刊介绍:
BBA Gene Regulatory Mechanisms includes reports that describe novel insights into mechanisms of transcriptional, post-transcriptional and translational gene regulation. Special emphasis is placed on papers that identify epigenetic mechanisms of gene regulation, including chromatin, modification, and remodeling. This section also encompasses mechanistic studies of regulatory proteins and protein complexes; regulatory or mechanistic aspects of RNA processing; regulation of expression by small RNAs; genomic analysis of gene expression patterns; and modeling of gene regulatory pathways. Papers describing gene promoters, enhancers, silencers or other regulatory DNA regions must incorporate significant functions studies.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


