A central research portal for mining pancreatic clinical and molecular datasets and accessing biobanked samples
IF 5
2区 医学
Q2 Medicine
引用次数: 0
Abstract
Objectives
We present Pancreas Genome Phenome Atlas (PGPA) as a resource for the mining and analysis of pancreatic -omics datasets, and demonstrate the biological interpretations possible due to this dynamic analytics hub accommodating an extensive range of publicly available datasets.
Methods
Clinical and molecular datasets from four primary sources are included (The Cancer Genome Atlas, International Cancer Genome Consortium, Cancer Cell Line Encyclopaedia, Genomics Evidence Neoplasia Information Exchange), which form the foundation of -omics profiling of pancreatic malignancies and related lesions (n = 7760 specimens). Several user-friendly analytical tools to integrate and explore molecular data derived from these primary specimens and cell lines are available. Crucially, PGPA is positioned as the data access point for Pancreatic Cancer Research Fund Tissue Bank – the only national pancreatic cancer biobank in the UK. This will pioneer a new era of biobanking to promote collaborative studies and effective sharing of multi-modal molecular, histopathology and imaging data (>125 000 specimens from >3980 cases and controls; >2700 radiology images, and >2630 digitised H&Es from 401 donors) to accelerate validation of in silico findings in patient-derived material.
Results
We demonstrate the practical utility of PGPA by investigating somatic variants associated with established transcriptomic subtypes and disease prognosis: several patient-specific variants are clinically actionable and may be leveraged for precision medicine.
Conclusions
This places PGPA at the analytical forefront of pancreatic biomarker-based research, providing the user community with a distinct resource to facilitate hypothesis-testing on public data, validate novel research findings, and access curated, high-quality patient tissues for translational research.
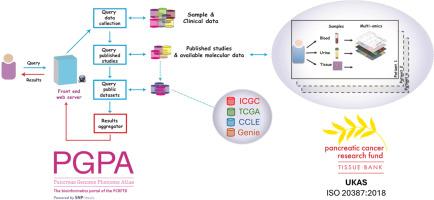
一个中央研究门户网站,用于挖掘胰腺临床和分子数据集,并访问生物库样本
目的:我们提出胰腺基因组表型图谱(PGPA)作为胰腺组学数据集挖掘和分析的资源,并展示由于这个动态分析中心容纳了广泛的公开可用数据集,可能的生物学解释。方法采用来自四个主要来源的临床和分子数据集(癌症基因组图谱、国际癌症基因组联盟、癌细胞系百科全书、基因组学证据肿瘤学信息交换),为胰腺恶性肿瘤及相关病变的组学分析奠定基础(n = 7760例标本)。一些用户友好的分析工具来整合和探索来自这些主要标本和细胞系的分子数据是可用的。至关重要的是,PGPA被定位为胰腺癌研究基金组织库(英国唯一的国家胰腺癌生物库)的数据访问点。这将开创一个生物银行的新时代,促进合作研究和多模式分子、组织病理学和成像数据的有效共享(来自3980例病例和对照组的125000个标本;2700个放射学图像;以及来自401个供体的2630个数字化H&; e),以加速对患者来源材料的计算机发现的验证。通过研究与已建立的转录组亚型和疾病预后相关的体细胞变异,我们证明了PGPA的实际效用:一些患者特异性变异在临床上是可行的,可能用于精准医学。这使得PGPA处于基于胰腺生物标志物研究的分析前沿,为用户社区提供了独特的资源,以促进对公共数据的假设检验,验证新的研究结果,并获得经过整理的高质量患者组织,用于转化研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Translational Oncology
ONCOLOGY-
CiteScore
8.40
自引率
2.00%
发文量
314
审稿时长
54 days
期刊介绍:
Translational Oncology publishes the results of novel research investigations which bridge the laboratory and clinical settings including risk assessment, cellular and molecular characterization, prevention, detection, diagnosis and treatment of human cancers with the overall goal of improving the clinical care of oncology patients. Translational Oncology will publish laboratory studies of novel therapeutic interventions as well as clinical trials which evaluate new treatment paradigms for cancer. Peer reviewed manuscript types include Original Reports, Reviews and Editorials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


