GRDGNN: A directed graph neural network framework for multi-relational inference of gene regulatory networks
IF 7.5
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Inferring gene regulatory networks (GRNs) from gene expression data is a central challenge in systems biology. Graph neural networks (GNNs) offer a promising approach due to their ability to process graph-structured data. However, existing GNN methods for GRN inference often treat the problem as binary classification, limiting their ability to capture comprehensive regulatory relationships. This paper introduces two learning algorithms that utilize an end-to-end gene regulatory directed graph neural network (GRDGNN) schema for efficient inference of causal relationships in large-scale networks. These algorithms incorporate a directed graph neural network (DGNN) and a graph multi-classification task to identify explicit interactions between transcription factors (TFs) and target genes. The proposed approach consists of four key steps: (1) constructing a directed initial network using regression Pearson correlation and mutual information analysis, (2) extracting subgraphs of observed TF-gene pairs and applying a DGNN for information aggregation, (3) projecting the aggregated information into a low-dimensional space using graph pooling to generate graph representations of TF-gene pairs, and (4) classifying subgraphs using a multilayer perceptron (MLP) for link prediction and inference of explicit regulatory relationships. Evaluation of the DREAM5 microarray and scRNA-seq datasets demonstrates that our transductive and inductive learning methods can accurately and effectively infer explicit regulatory relationships compared to benchmark methods. These results demonstrate that the proposed GRDGNN schema exhibits strong generalization across species, data types, and modalities in cross-species, cross-data type, and cross-modality learning.
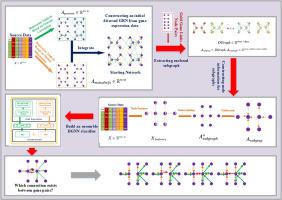
GRDGNN:用于基因调控网络多关系推理的有向图神经网络框架
从基因表达数据推断基因调控网络(grn)是系统生物学的核心挑战。图神经网络(gnn)由于其处理图结构数据的能力而提供了一种很有前途的方法。然而,现有的用于GRN推理的GNN方法通常将问题视为二元分类,限制了它们捕捉全面调节关系的能力。本文介绍了两种学习算法,它们利用端到端基因调控有向图神经网络(GRDGNN)模式对大规模网络中的因果关系进行有效推断。这些算法结合了有向图神经网络(DGNN)和图多分类任务来识别转录因子(tf)和靶基因之间的显式相互作用。拟议的办法包括四个关键步骤:(1)利用回归Pearson相关和互信息分析构建有向初始网络;(2)提取观察到的tf基因对的子图,并应用DGNN进行信息聚合;(3)利用图池将聚合信息投影到低维空间,生成tf基因对的图表示;(4)利用多层感知器(MLP)对子图进行分类,进行链路预测和显性调控关系推理。对DREAM5微阵列和scRNA-seq数据集的评估表明,与基准方法相比,我们的传导和归纳学习方法可以准确有效地推断明确的调控关系。这些结果表明,所提出的GRDGNN模式在跨物种、跨数据类型和跨模态学习中具有很强的跨物种、数据类型和模态的泛化能力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Expert Systems with Applications
工程技术-工程:电子与电气
CiteScore
13.80
自引率
10.60%
发文量
2045
审稿时长
8.7 months
期刊介绍:
Expert Systems With Applications is an international journal dedicated to the exchange of information on expert and intelligent systems used globally in industry, government, and universities. The journal emphasizes original papers covering the design, development, testing, implementation, and management of these systems, offering practical guidelines. It spans various sectors such as finance, engineering, marketing, law, project management, information management, medicine, and more. The journal also welcomes papers on multi-agent systems, knowledge management, neural networks, knowledge discovery, data mining, and other related areas, excluding applications to military/defense systems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


