Investigating the molecular basis of cleptobiosis in eusocial stingless bees (Apidae: Hymenoptera)
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2025-09-28
DOI:10.1016/j.cbd.2025.101645
引用次数: 0
Abstract
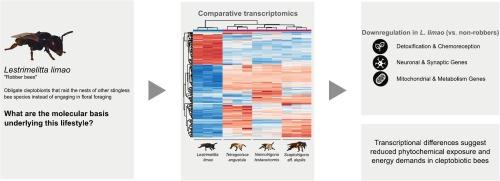
Cleptobiosis, the act of raiding other species to obtain food or resources, is widespread among animals but rarely obligatory. The eusocial stingless bee Lestrimelitta limao is one of the few species that depend entirely on cleptobiosis, raiding other stingless bee colonies for survival. To investigate the molecular basis and putative evolutionary adaptations associated with this specialized lifestyle, we compared the transcriptomes of non-raiding foragers of L. limao and of three non-robber stingless bees – Nannotrigona testaceicornis, Scaptotrigona aff. depilis, and Tetragonisca angustula.
Our analysis revealed that differentially expressed orthologs were predominantly downregulated in L. limao workers, suggesting reduced transcriptional activity during foraging in this species. These downregulated genes fall into three major functional categories potentially linked to cleptobiotic adaptations: (1) detoxification and chemoreception genes, including cytochrome P450s and odorant receptors, indicating decreased exposure to phytochemicals; (2) neuronal and synaptic genes, such as para and Dys, possibly reflecting neurophysiological modifications; and (3) mitochondrial and carbohydrate metabolism genes, suggesting lower energetic demands. These findings provide novel insights into the molecular mechanisms associated with cleptobiotic behavior in eusocial bees.
调查的分子基础cleptobiosis在群居蜜蜂无刺的膜翅目昆虫(蜜蜂科:)
掠食是一种掠食其他物种以获取食物或资源的行为,在动物中很普遍,但很少是强制性的。群居无刺蜜蜂Lestrimelitta limmao是少数完全依赖钩端寄生的物种之一,它们会袭击其他无刺蜜蜂群体以生存。为了研究这种特殊生活方式的分子基础和可能的进化适应性,我们比较了非掠食觅食蜜蜂L. limao和三种非掠食无刺蜜蜂——Nannotrigona testaceicornis、Scaptotrigona affs . depilis和Tetragonisca angustula的转录组。我们的分析显示,差异表达的同源物在L. limao工蜂中主要下调,表明该物种在觅食过程中转录活性降低。这些下调的基因分为三大类,可能与生物适应有关:(1)解毒和化学接受基因,包括细胞色素p450和气味受体,表明暴露于植物化学物质的减少;(2)神经元和突触基因,如para和Dys,可能反映神经生理改变;(3)线粒体和碳水化合物代谢基因,表明较低的能量需求。这些发现提供了新的见解与分子机制相关的钩生行为在群居蜜蜂。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


