Correlated latent space learning for structural differentiation modeling in single cell RNA data
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
Single-cell RNA sequencing (scRNA-seq) enables high-resolution analysis of cellular differentiation, yet many existing methods have difficulty modeling its continuous, coupled, and noise-prone dynamics. We present CODEVAE (Correlated Ordinary Differential Equation Variational Autoencoder), a deep generative framework that integrates ordinary differential equation constraints with correlation-aware latent representations to preserve geometric continuity and biologically coupled variation. Building on a baseline variational autoencoder, CODEVAE incrementally incorporates low- regularization, an information bottleneck reconstruction pathway, ODE-based continuity, and correlated latent components. Across an evaluation suite of 18 metrics and 55 independent runs, CODEVAE achieves consistently higher performance than advanced variational models, single-cell specific methods, graph/contrastive approaches, and traditional dimensionality reduction techniques. In multi-batch settings, CODEVAE maintains smooth manifolds and attains improved integration quality. In biological applications, CODEVAE reconstructs a continuous megakaryocyte differentiation trajectory and delineates stage-specific effects of Dapp1 perturbation. These findings position CODEVAE as a robust, principled approach for modeling continuous cellular dynamics and extracting mechanistic insights across diverse single-cell contexts.
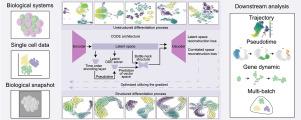
单细胞RNA数据中结构分化建模的相关潜在空间学习
单细胞RNA测序(scRNA-seq)能够对细胞分化进行高分辨率分析,但许多现有方法难以对其连续、耦合和易受噪声影响的动态建模。我们提出了CODEVAE(相关常微分方程变分自编码器),这是一个深度生成框架,它将常微分方程约束与相关感知的潜在表示集成在一起,以保持几何连续性和生物耦合变化。CODEVAE以基线变分自编码器为基础,逐步整合了低β正则化、信息瓶颈重构路径、基于ode的连续性和相关潜在分量。在18个指标和55个独立运行的评估套件中,CODEVAE实现了比高级变分模型、单细胞特定方法、图/对比方法和传统降维技术始终更高的性能。在多批设置中,CODEVAE保持光滑的流形并实现改进的集成质量。在生物学应用中,CODEVAE重建了一个连续的巨核细胞分化轨迹,并描绘了Dapp1扰动的阶段特异性效应。这些发现将CODEVAE定位为一种强大的、有原则的方法,用于建模连续的细胞动力学,并在不同的单细胞环境中提取机制见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


