PARP1 auto-modification promotes faithful Okazaki fragment processing and limits replication fork speed
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Poly(ADP-ribose) polymerase (PARP) inhibitors have proven their efficacy for treating tumors defective in homologous recombination via synthetic lethality. In response to DNA breaks, PARP1 is the primary ADP-ribosylation writer, modifying itself (auto-modification) and other proteins to facilitate repair. However, enzymatic inhibition blocks both processes, making it difficult to dissect their distinct functional roles. Using proteomics and site-directed mutagenesis, we identified a PARP1 mutant deficient in auto-modification, yet it retains catalytic activity. This separation-of-function mutant revealed that PARP1 auto-modification slows DNA replication fork progression but is dispensable for repair factor recruitment. Instead, auto-modification promotes the timely release of PARP1 at DNA break sites and prevents the formation of replication stress. Simultaneous inhibition of FEN1 and loss of PARP1 auto-modification gives rise to synthetic lethality, implicating auto-modification in Okazaki fragment processing. Our results demonstrate that trapping of PARP at DNA breaks impedes repair factor accessibility, constituting an important dimension of PARP-inhibitor-driven cytotoxicity.
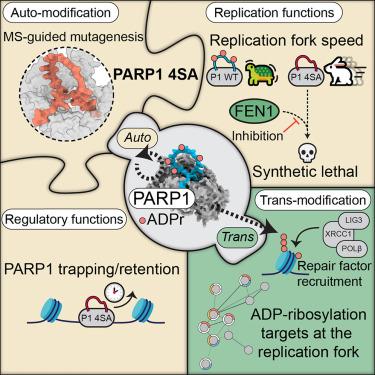
PARP1的自动修改促进了忠实的冈崎片段处理,并限制了复制叉的速度
聚(adp -核糖)聚合酶(PARP)抑制剂通过合成致死性治疗同源重组有缺陷的肿瘤已被证明有疗效。在DNA断裂的反应中,PARP1是主要的adp -核糖基化书写者,修饰自身(自修饰)和其他蛋白质以促进修复。然而,酶抑制阻断了这两个过程,使得很难解剖它们不同的功能角色。利用蛋白质组学和定点诱变技术,我们发现了一个PARP1突变体缺乏自修饰,但它保持了催化活性。这种功能分离突变表明,PARP1的自修饰减缓了DNA复制叉的进展,但对于修复因子的募集是必不可少的。相反,自修饰促进PARP1在DNA断裂位点的及时释放,防止复制应激的形成。同时抑制FEN1和丧失PARP1自修饰可导致合成致死性,暗示了冈崎片段加工中的自修饰。我们的研究结果表明,PARP在DNA断裂处的捕获阻碍了修复因子的可及性,这是PARP抑制剂驱动的细胞毒性的一个重要方面。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


