Quantum LBP-driven heart sound analysis with quality assessment in real-world noisy environments
IF 4.8
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background and Objective:
Cardiovascular disease (CVD) is a major global health concern with increasing prevalence. While electrocardiography (ECG) and echocardiography offer high accuracy, their reliance on specialized equipment and trained personnel makes them costly and less accessible in rural areas. In contrast, the Phonocardiography (PCG) provides a more affordable alternative via capturing heart sound signals using a stethoscope. However, PCG analysis is often compromised by environmental noise. Most existing methods address this issue with denoising techniques, which can inadvertently suppress vital heart sound components. To address this challenge, the objective of this work is to develop a computationally efficient and noise-resilient method for PCG classification.
Methods:
The proposed approach introduces a quality assessment metric () to select the least noisy subsequences for further processing. For noise-robust feature extraction, an Enhanced Quantum Local Binary Pattern (EQLBP) method is employed, which adaptively selects reference pixels and extracts uniform patterns from the signal spectrogram to mitigate noise effects. In addition, Discrete Wavelet Transform (DWT) features are extracted to capture multi-resolution time–frequency characteristics, complementing the local texture features obtained from EQLBP. The combined feature set is then used to train conventional machine learning models.
Results:
The proposed method was evaluated using 10-fold cross-validation on two publicly available datasets: CinC-2016 and HSM-2018. On CinC-2016, it achieved an accuracy of 97.22%, precision of 98.29%, recall of 98.63%, and an F1-score of 98.46%. On HSM-2018, the method obtained an accuracy of 98.70%, precision of 99.05%, recall of 99.00%, and an F1-score of 99.00%. These results highlight the superior performance of the proposed approach compared to existing methods.
Conclusions:
With its computational efficiency and robust performance, the proposed method is well-suited for out-of-clinic applications, particularly in rural and remote areas where access to advanced diagnostic tools is limited.
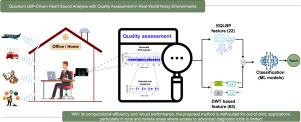
量子lbp驱动的心音分析与现实世界嘈杂环境中的质量评估
背景与目的:心血管疾病(CVD)是一个主要的全球健康问题,患病率不断上升。虽然心电图(ECG)和超声心动图提供了很高的准确性,但它们对专业设备和训练有素的人员的依赖使得它们成本高昂,而且在农村地区很难获得。相比之下,心音图(PCG)通过使用听诊器捕捉心音信号,提供了一种更实惠的替代方案。然而,PCG分析经常受到环境噪声的影响。大多数现有的方法都是用去噪技术来解决这个问题,这可能会无意中抑制重要的心音成分。为了应对这一挑战,本工作的目标是开发一种计算效率高、抗噪声的PCG分类方法。方法:该方法引入质量评估度量(PCGQA)来选择噪声最小的子序列进行进一步处理。在抗噪声特征提取方面,采用增强量子局部二值模式(Enhanced Quantum Local Binary Pattern, EQLBP)方法,自适应地选择参考像素点,从信号谱图中提取均匀模式,以减轻噪声影响。此外,提取离散小波变换(DWT)特征,捕获多分辨率时频特征,补充EQLBP获得的局部纹理特征。然后使用组合的特征集来训练传统的机器学习模型。结果:在两个公开可用的数据集(cdc -2016和HSM-2018)上使用10倍交叉验证对所提出的方法进行了评估。在cic -2016上,准确率为97.22%,精密度为98.29%,召回率为98.63%,f1得分为98.46%。在HSM-2018上,该方法的准确率为98.70%,精密度为99.05%,召回率为99.00%,f1评分为99.00%。这些结果突出了该方法与现有方法相比的优越性能。结论:该方法计算效率高,性能稳健,非常适合临床外应用,特别是在农村和偏远地区,在那里获得先进的诊断工具是有限的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


