Identification of novel selective estrogen receptor degraders (SERD) via physics-based and deep-learning-based virtual screening and Bioassys
IF 4.7
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Breast cancer is the most common malignant tumor among women, most of which are ERα(Estrogen Receptor alpha) positive. SERDs(Selective Estrogen Receptor Degraders), such as Fulvestrant(the first SERD), can induce degradation of this receptor, leading to overcome the acquired endocrine resistance. However, only two SERDs (Fulvestrant and Elacestrant) are currently clinically approved, highlighting an urgent demand for novel, oral, and more potent alternatives. In this study we developed a multi-tiered virtual screening combing physics-based docking methods (Glide) with deep-learning-based docking methods (Karmadock and Carsidock) to identify SERDs with novel scaffold. After ADMET and MM-GBSA screening, four purchasable candidate compounds were selected for biological evaluation in three cell lines. Among them, two compounds exhibited significant anti-proliferation activity aganist ER-positive cells. The fingerprint analysis also revealed their structural novelty, which are distinct from the known SERDs. Further study indicated F0840–0093 could directly bound to ERα and induced its proteasomal degradation (mimicking Fulvestrant). In summary, our work not only provided a feasible virtual screening approach in drug discovery but also identified some compounds, particularly F0840–0093, which can be a promising lead with new chemical scaffold for further optimization and development as SERDs.
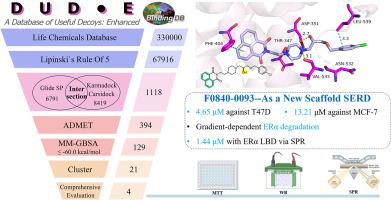
通过基于物理和深度学习的虚拟筛选和Bioassys鉴定新型选择性雌激素受体降解物(SERD)
乳腺癌是女性中最常见的恶性肿瘤,多数为ERα(雌激素受体α)阳性。选择性雌激素受体降解剂(SERD),如氟维司汀(第一种SERD),可以诱导该受体的降解,从而克服获得性内分泌抵抗。然而,目前只有两种serd (Fulvestrant和Elacestrant)获得临床批准,这凸显了对新型、口服和更有效替代品的迫切需求。在这项研究中,我们开发了一种多层虚拟筛选方法,结合基于物理的对接方法(Glide)和基于深度学习的对接方法(Karmadock和Carsidock)来识别具有新型支架的serd。经过ADMET和MM-GBSA筛选,选择四种可购买的候选化合物在三种细胞系中进行生物学评价。其中,两种化合物对er阳性细胞具有显著的抗增殖活性。指纹图谱分析还揭示了它们的结构新颖性,与已知的serd不同。进一步研究表明,F0840-0093可以直接结合ERα并诱导其蛋白酶体降解(类似于氟维司坦)。综上所述,我们的工作不仅为药物发现提供了一种可行的虚拟筛选方法,而且还发现了一些化合物,特别是F0840-0093,可以作为新的化学支架进一步优化和开发serd的有希望的线索。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bioorganic Chemistry
生物-生化与分子生物学
CiteScore
9.70
自引率
3.90%
发文量
679
审稿时长
31 days
期刊介绍:
Bioorganic Chemistry publishes research that addresses biological questions at the molecular level, using organic chemistry and principles of physical organic chemistry. The scope of the journal covers a range of topics at the organic chemistry-biology interface, including: enzyme catalysis, biotransformation and enzyme inhibition; nucleic acids chemistry; medicinal chemistry; natural product chemistry, natural product synthesis and natural product biosynthesis; antimicrobial agents; lipid and peptide chemistry; biophysical chemistry; biological probes; bio-orthogonal chemistry and biomimetic chemistry.
For manuscripts dealing with synthetic bioactive compounds, the Journal requires that the molecular target of the compounds described must be known, and must be demonstrated experimentally in the manuscript. For studies involving natural products, if the molecular target is unknown, some data beyond simple cell-based toxicity studies to provide insight into the mechanism of action is required. Studies supported by molecular docking are welcome, but must be supported by experimental data. The Journal does not consider manuscripts that are purely theoretical or computational in nature.
The Journal publishes regular articles, short communications and reviews. Reviews are normally invited by Editors or Editorial Board members. Authors of unsolicited reviews should first contact an Editor or Editorial Board member to determine whether the proposed article is within the scope of the Journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


