Multimodal profiling of Pepcan-CB1 receptor structure-activity relationships: integrating molecular dynamics simulations, biological profiling, and the deep learning model MuMoPepcan
IF 4.7
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
In machine learning of drug discovery, the scale of accessible data is often strictly limited, while few-shot learning in wet-lab experimental data limits the accuracy of machine learning algorithms. Cannabinoid receptors are involved in various important physiological activities, and pepcans are key components of the endocannabinoid system. Herein, we proposed a combined dry-wet lab experimental framework that incorporated molecular dynamics simulation (MDS) data into peptide biological activity prediction. We validated our hypothesis on cannabinoid receptors type 1 (CB1) and pepcans: (1) In the study, we synthesized 45 pepcan peptides to establish a bioactivity dataset and identified RD-pepcan-11 as an lead analgesic compound by Bioscreening, with systematic characterization of its CB1 selectivity and pharmacodynamics.; (2) Millions of conformational data were generated by MDS and a CB1-pepcans conformation dataset was constructed; (3) Combining wet-lab data and MDS data, a deep learning model - MuMoPepcan was developed, reducing prediction errors to within the error range of wet-lab experiments. This study not only identified novel high-potential pepcans - RD-pepcan-11, but also demonstrated that MDS can serve as an effective data augmentation method to scale up drug-receptor datasets, thereby improving model generalizability and performance.
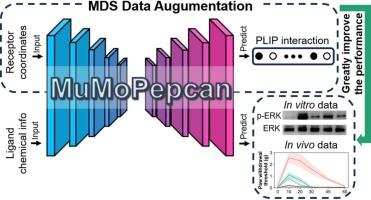
Pepcan-CB1受体结构-活性关系的多模态分析:整合分子动力学模拟、生物学分析和深度学习模型MuMoPepcan
在药物发现的机器学习中,可访问数据的规模往往受到严格限制,而湿实验室实验数据中的少量学习限制了机器学习算法的准确性。大麻素受体参与多种重要的生理活动,而胃管是内源性大麻素系统的关键组成部分。在此,我们提出了一个干湿联合实验室实验框架,将分子动力学模拟(MDS)数据纳入肽生物活性预测。我们验证了我们对大麻素受体1型(CB1)和肽的假设:(1)在研究中,我们合成了45个肽,建立了肽的生物活性数据集,并通过生物筛选鉴定了RD-pepcan-11为先导镇痛化合物,并对其CB1选择性和药效学进行了系统表征;(2) MDS生成了数百万个构象数据,构建了CB1-pepcans构象数据集;(3)结合湿实验室数据和MDS数据,开发了一种深度学习模型——MuMoPepcan,将预测误差降低到湿实验室实验误差范围内。本研究不仅鉴定出了新型的高电位pepcan- RD-pepcan-11,而且还证明了MDS可以作为一种有效的数据增强方法来扩大药物受体数据集,从而提高模型的泛化性和性能。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bioorganic Chemistry
生物-生化与分子生物学
CiteScore
9.70
自引率
3.90%
发文量
679
审稿时长
31 days
期刊介绍:
Bioorganic Chemistry publishes research that addresses biological questions at the molecular level, using organic chemistry and principles of physical organic chemistry. The scope of the journal covers a range of topics at the organic chemistry-biology interface, including: enzyme catalysis, biotransformation and enzyme inhibition; nucleic acids chemistry; medicinal chemistry; natural product chemistry, natural product synthesis and natural product biosynthesis; antimicrobial agents; lipid and peptide chemistry; biophysical chemistry; biological probes; bio-orthogonal chemistry and biomimetic chemistry.
For manuscripts dealing with synthetic bioactive compounds, the Journal requires that the molecular target of the compounds described must be known, and must be demonstrated experimentally in the manuscript. For studies involving natural products, if the molecular target is unknown, some data beyond simple cell-based toxicity studies to provide insight into the mechanism of action is required. Studies supported by molecular docking are welcome, but must be supported by experimental data. The Journal does not consider manuscripts that are purely theoretical or computational in nature.
The Journal publishes regular articles, short communications and reviews. Reviews are normally invited by Editors or Editorial Board members. Authors of unsolicited reviews should first contact an Editor or Editorial Board member to determine whether the proposed article is within the scope of the Journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


