Precise ligand-selective mechanism at the fab domain of a tau-recognizing antibody
IF 2.7
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
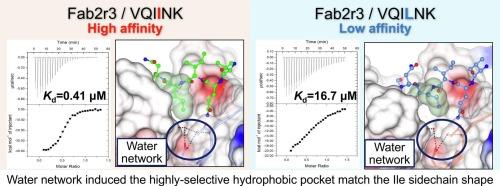
Insoluble aggregated tau protein in the form of paired helical filaments is a causative agent of the neurofibrillary tangles observed in Alzheimer’s disease (AD). The hexapeptide 275VQIINK280 located in the microtubule-binding domain of tau plays a crucial role in the abnormal aggregation process. Therefore, targeting the VQIINK sequence with a tau aggregation inhibitor may be a promising therapeutic approach for AD. A previous study demonstrated that the Fab domain of the tau antibody (Fab2r3) inhibits tau aggregation by binding to the VQIINK sequence. By determining the three-dimensional structures of the Fab2r3-VQIINK peptide complex and apo Fab2r3, we elucidated the recognition mechanism between Fab2r3 and the VQIINK peptide. However, the basis for the selectivity of Fab2r3 for VQIINK was not completely clear. Therefore, the objective of this report is to investigate the selective binding mechanism of Fab2r3 against VQIINK peptide. Through isothermal titration calorimetry, we show that Ile-4 in the VQIINK peptide is crucial for the selectivity of Fab2r3. X-ray structural analysis of three complexes of Fab2r3 with Ile-4 mutated peptides (VQIVYK, VQILNK, and VQIFNK) suggested that the rigid conformation of a hydrophobic pocket in Fab2r3 plays a vital role in ligand selectivity. These findings may explain the effectiveness of Fab2r3 as a tau aggregation inhibitor.
tau识别抗体fab结构域的精确配体选择机制。
成对螺旋细丝形式的不溶性聚集tau蛋白是阿尔茨海默病(AD)中观察到的神经原纤维缠结的病原体。位于tau蛋白微管结合区域的六肽275VQIINK280在异常聚集过程中起着至关重要的作用。因此,用tau聚集抑制剂靶向VQIINK序列可能是一种很有前景的治疗AD的方法。先前的研究表明,tau抗体(Fab2r3)的Fab结构域通过结合VQIINK序列抑制tau聚集。通过测定Fab2r3-VQIINK肽复合物和载子Fab2r3的三维结构,我们阐明了Fab2r3与VQIINK肽之间的识别机制。然而,Fab2r3对VQIINK选择性的基础并不完全清楚。因此,本报告的目的是研究Fab2r3对VQIINK肽的选择性结合机制。通过等温滴定量热法,我们发现VQIINK肽中的Ile-4对Fab2r3的选择性至关重要。对三种与Ile-4突变肽(VQIVYK、VQILNK和VQIFNK)的Fab2r3配合物的x射线结构分析表明,Fab2r3疏水袋的刚性构象对配体选择性起着至关重要的作用。这些发现可以解释Fab2r3作为tau聚集抑制剂的有效性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of structural biology
生物-生化与分子生物学
CiteScore
6.30
自引率
3.30%
发文量
88
审稿时长
65 days
期刊介绍:
Journal of Structural Biology (JSB) has an open access mirror journal, the Journal of Structural Biology: X (JSBX), sharing the same aims and scope, editorial team, submission system and rigorous peer review. Since both journals share the same editorial system, you may submit your manuscript via either journal homepage. You will be prompted during submission (and revision) to choose in which to publish your article. The editors and reviewers are not aware of the choice you made until the article has been published online. JSB and JSBX publish papers dealing with the structural analysis of living material at every level of organization by all methods that lead to an understanding of biological function in terms of molecular and supermolecular structure.
Techniques covered include:
• Light microscopy including confocal microscopy
• All types of electron microscopy
• X-ray diffraction
• Nuclear magnetic resonance
• Scanning force microscopy, scanning probe microscopy, and tunneling microscopy
• Digital image processing
• Computational insights into structure
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


