TRIM52 Ubiquitin Ligase Acts as a Key Recognition Component of the Mammalian fMet/N-degron Pathway
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Eukaryotic translation can initiate with formylmethionine (fMet), generating N-terminally formylated proteins in mitochondria and, unexpectedly, in the cytosol. However, the specific mechanism for eliminating cytosolic fMet-bearing proteins has remained elusive. Here, we identify the E3 ubiquitin ligase TRIM52 as the key recognition component of the mammalian fMet/N-degron pathway. TRIM52 targets N-terminally formylated proteins, including TPD54 and SPTAN1, for proteasomal degradation. It recognizes the N-terminal fMet through an evolutionarily conserved acidic loop embedded between its bipartite RING domain. Structural modeling and mutagenesis identify Tyr148 in the acidic loop as critical for fMet recognition without intervening E3 ligase activity. TRIM52 depletion stabilizes fMet-bearing proteins, disrupts proteostasis, and induces caspase-3-dependent apoptosis—phenotypes rescued by enhancing deformylation. These findings establish TRIM52 as a dedicated sensor and effector of the mammalian fMet/N-degron pathway, linking N-terminal formylation to proteostasis and cell survival.
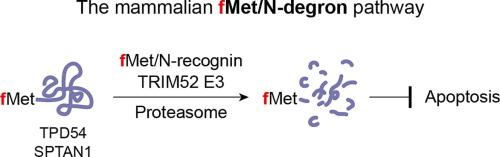
TRIM52泛素连接酶是哺乳动物fMet/N-degron通路的关键识别组分。
真核翻译可以由甲酰基蛋氨酸(fMet)启动,在线粒体中产生n端甲酰基化蛋白,并且意外地在细胞质中产生。然而,消除细胞质中含fmet蛋白的具体机制仍然难以捉摸。在这里,我们发现E3泛素连接酶TRIM52是哺乳动物fMet/N-degron通路的关键识别组分。TRIM52靶向n端甲酰化蛋白,包括TPD54和SPTAN1,用于蛋白酶体降解。它通过嵌入在其两部分环结构域之间的进化保守的酸性环识别n端fMet。结构建模和诱变发现,在不干预E3连接酶活性的情况下,酸性环中的Tyr148对fMet识别至关重要。TRIM52的缺失稳定了携带fmet的蛋白,破坏了蛋白质稳态,并诱导caspase-3依赖性凋亡-通过增强去甲酰基化来拯救表型。这些发现表明TRIM52是哺乳动物fMet/N-degron通路的专用传感器和效应器,将n端甲酰化与蛋白质静止和细胞存活联系起来。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


