Unraveling the Diversity of RANBP2: Protein Isoforms and Implications for Cellular Function and Human Disease
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
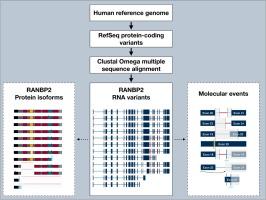
RANBP2 (also known as Nup358) is the largest nucleoporin of the nuclear pore complex (NPC), where it constitutes the main component of the cytoplasmic filaments and regulates nucleocytoplasmic transport. In addition to its NPC-associated functions, RANBP2 also localizes in the endoplasmic reticulum (ER) at annulate lamellae (AL), at mitochondria-ER junctions, and at kinetochores during mitosis, where it contributes to multiple cellular processes including metabolism and mitotic progression. Although most studies focus on the canonical full-length protein (∼358 kDa in humans), multiple smaller bands have been detected across species, suggesting the existence of alternative isoforms. Here, we review all predicted and experimentally supported RANBP2 transcript variants, summarize their structural features and discuss their possible origins, including alternative splicing, genomic recombination and proteolysis. We examine how isoform-specific changes, such as loss of the zinc finger domain, Ran-binding domains, E3 SUMO ligase, or the cyclophilin-like domain, could alter RANBP2′s cellular functions. We also consider evidence for cell-type specific and developmentally regulated expression from non-human models, and evaluate the potential relevance of RANBP2 isoforms in viral infections and neurological disease. By compiling genomic, proteomic, and functional data, this review highlights the need for isoform-resolved approaches to fully understand RANBP2 biology and its contribution to human disease.
揭示RANBP2的多样性:蛋白质亚型及其对细胞功能和人类疾病的影响。
RANBP2(也被称为Nup358)是核孔复合物(NPC)中最大的核孔蛋白,它构成细胞质丝的主要成分,并调节核胞质运输。除了npc相关功能外,RANBP2还定位于内质网(ER)环状片层(AL)、线粒体-内质网连接处和有丝分裂过程中的着丝点,在那里它参与多种细胞过程,包括代谢和有丝分裂过程。尽管大多数研究集中在典型的全长蛋白(人类中约358 kDa)上,但在不同物种中发现了多个更小的条带,这表明存在其他同种异构体。在这里,我们回顾了所有预测和实验支持的RANBP2转录变体,总结了它们的结构特征,并讨论了它们可能的起源,包括选择性剪接、基因组重组和蛋白质水解。我们研究了异构体特异性变化,如锌指结构域、ran结合结构域、E3 SUMO连接酶或亲环蛋白样结构域的缺失,如何改变RANBP2的细胞功能。我们还考虑了来自非人类模型的细胞类型特异性和发育调节表达的证据,并评估了RANBP2亚型在病毒感染和神经系统疾病中的潜在相关性。通过编译基因组学、蛋白质组学和功能数据,本综述强调了对同种异构体解析方法的需求,以充分了解RANBP2生物学及其对人类疾病的贡献。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


