Identification of a potential anti-nontuberculous mycobacterial drug candidate targeting a mycothiol disulfide reductase
IF 4.7
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The resistance of nontuberculous mycobacteria (NTM) to conventional anti-tuberculosis drugs and its growing infection rate year by year urgently require new treatment strategies. Structure-based virtual screening, which can greatly improve efficiency and reduce costs in the early stage of drug development, is an indispensable part of modern drug discovery. In this study, the crystal structure of the mycothiol disulfide reductase from Mycobacterium abscessus (MabMtr) was determined. Through virtual screening, compound AK-968/11492032 was identified as a promising candidate capable of fitting well into the potential MSSM-binding pocket of MabMtr. It was discovered that AK-968/11492032 and its derivatives (Y6B and Y6C) could produce antimicrobial effects on the Mycobacterial type strain Mycobacterium smegmatis. Moreover, microscale thermophoresis analysis was employed to evaluate the high binding affinity of the compounds to MabMtr. Furthermore, the key residues (S14, I47, H451) of MabMtr involved in the interaction with AK-968/11492032 were predicted and confirmed through molecular docking and mutational analysis, MabMtr was verified as the target for it to exert antibacterial effects through in vitro enzyme activity and in vivo gene knockout, complementation, and overexpression. These findings provide a potential development target to develop effective and specific anti-NTM drugs.
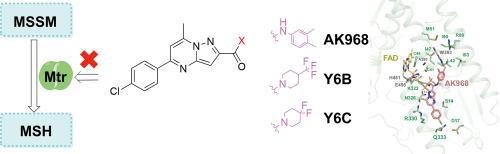
一种潜在的抗非结核分枝杆菌候选药物的鉴定,靶向一种菌硫醇二硫还原酶。
非结核分枝杆菌(NTM)对常规抗结核药物的耐药性及其逐年上升的感染率迫切需要新的治疗策略。基于结构的虚拟筛选可以在药物开发的早期阶段大大提高效率和降低成本,是现代药物发现不可缺少的一部分。本研究测定了脓肿分枝杆菌(Mycobacterium abessus, MabMtr)中菌硫醇二硫还原酶(mycothiol二硫还原酶)的晶体结构。通过虚拟筛选,化合物AK-968/11492032被确定为有希望的候选物,能够很好地适应MabMtr潜在的mssm结合口袋。发现AK-968/11492032及其衍生物Y6B和Y6C对耻垢分枝杆菌型菌株有抗菌作用。此外,采用微尺度热泳分析评价了化合物与MabMtr的高结合亲和力。通过分子对接和突变分析,预测并确认了MabMtr与AK-968/11492032相互作用的关键残基(S14、I47、H451),通过体外酶活性和体内基因敲除、互补、过表达验证了MabMtr是其发挥抗菌作用的靶点。这些发现为开发有效的特异性抗ntm药物提供了一个潜在的开发靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bioorganic Chemistry
生物-生化与分子生物学
CiteScore
9.70
自引率
3.90%
发文量
679
审稿时长
31 days
期刊介绍:
Bioorganic Chemistry publishes research that addresses biological questions at the molecular level, using organic chemistry and principles of physical organic chemistry. The scope of the journal covers a range of topics at the organic chemistry-biology interface, including: enzyme catalysis, biotransformation and enzyme inhibition; nucleic acids chemistry; medicinal chemistry; natural product chemistry, natural product synthesis and natural product biosynthesis; antimicrobial agents; lipid and peptide chemistry; biophysical chemistry; biological probes; bio-orthogonal chemistry and biomimetic chemistry.
For manuscripts dealing with synthetic bioactive compounds, the Journal requires that the molecular target of the compounds described must be known, and must be demonstrated experimentally in the manuscript. For studies involving natural products, if the molecular target is unknown, some data beyond simple cell-based toxicity studies to provide insight into the mechanism of action is required. Studies supported by molecular docking are welcome, but must be supported by experimental data. The Journal does not consider manuscripts that are purely theoretical or computational in nature.
The Journal publishes regular articles, short communications and reviews. Reviews are normally invited by Editors or Editorial Board members. Authors of unsolicited reviews should first contact an Editor or Editorial Board member to determine whether the proposed article is within the scope of the Journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


