Resistance mutations, drug binding and drug residence times
IF 6.1
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The rapid evolution of microorganisms and cancer cells makes it difficult to treat tumours and infectious diseases, because resistance to drugs is the rule rather than the exception. Structures or models of protein–drug complexes help to understand how mutations lead to resistance and to design better drugs. However, it is difficult to reason how small changes in the structure lead to drug resistance. Thus, protein and drug dynamics need to be considered. Strategies to increase drug residence are sought after to increase the efficacy of drugs. Computational methods to calculate the effect of mutations on drug binding and residence times are being developed and improved, but are challenging. A priori prediction of a mutation's effect on drug binding is an even greater challenge. On the other hand, knowledge about protein–drug complexes has led to the development of multiple design strategies that aim to reduce mutation-driven drug resistance.
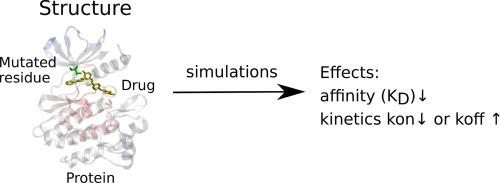
耐药突变、药物结合和药物停留时间。
微生物和癌细胞的快速进化使肿瘤和传染病的治疗变得困难,因为对药物的耐药性是普遍现象,而不是例外。蛋白质-药物复合物的结构或模型有助于了解突变如何导致耐药性和设计更好的药物。然而,很难解释结构上的微小变化是如何导致耐药性的。因此,需要考虑蛋白质和药物动力学。为了提高药物的疗效,人们一直在寻求增加药物驻留的策略。计算突变对药物结合和停留时间影响的计算方法正在发展和改进,但具有挑战性。先验地预测突变对药物结合的影响是一个更大的挑战。另一方面,关于蛋白质-药物复合物的知识导致了多种设计策略的发展,旨在减少突变驱动的耐药性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current opinion in structural biology
生物-生化与分子生物学
CiteScore
12.20
自引率
2.90%
发文量
179
审稿时长
6-12 weeks
期刊介绍:
Current Opinion in Structural Biology (COSB) aims to stimulate scientifically grounded, interdisciplinary, multi-scale debate and exchange of ideas. It contains polished, concise and timely reviews and opinions, with particular emphasis on those articles published in the past two years. In addition to describing recent trends, the authors are encouraged to give their subjective opinion of the topics discussed.
In COSB, we help the reader by providing in a systematic manner:
1. The views of experts on current advances in their field in a clear and readable form.
2. Evaluations of the most interesting papers, annotated by experts, from the great wealth of original publications.
[...]
The subject of Structural Biology is divided into twelve themed sections, each of which is reviewed once a year. Each issue contains two sections, and the amount of space devoted to each section is related to its importance.
-Folding and Binding-
Nucleic acids and their protein complexes-
Macromolecular Machines-
Theory and Simulation-
Sequences and Topology-
New constructs and expression of proteins-
Membranes-
Engineering and Design-
Carbohydrate-protein interactions and glycosylation-
Biophysical and molecular biological methods-
Multi-protein assemblies in signalling-
Catalysis and Regulation
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


