From snapshots to ensembles: Integrating experimental data and dynamics
IF 6.1
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
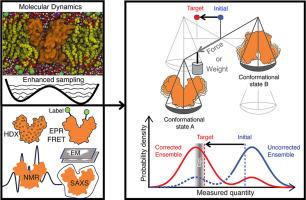
Protein function arises from the interplay of structure, dynamics, and biomolecular interactions. Despite advances in cryo-EM and AI-based structure prediction, capturing dynamic and energetic features remains a challenge. Biophysical methods like NMR, EPR, HDX-MS, SAXS, and cryo-EM provide valuable but often indirect signals. Connecting these to molecular mechanisms requires integrative approaches that combine experiments with physics-based simulations, revealing both stable structures and transient, functionally important intermediates. This review highlights recent advances in integrative modeling using the maximum entropy principle to build dynamic ensembles from diverse data while addressing uncertainty and bias. These methods help resolve heterogeneity and interpret low-resolution data. We conclude by exploring how integrative modeling, enhanced sampling, and AI-driven tools enable new insights into slow, large-scale conformational changes.
从快照到集成:整合实验数据和动力学
蛋白质的功能源于结构、动力学和生物分子相互作用的相互作用。尽管低温电镜和基于人工智能的结构预测取得了进展,但捕捉动态和能量特征仍然是一个挑战。生物物理方法如NMR, EPR, HDX-MS, SAXS和cro - em提供有价值但通常是间接的信号。将这些与分子机制联系起来需要综合的方法,将实验与基于物理的模拟相结合,揭示稳定的结构和瞬态的,功能重要的中间体。这篇综述强调了综合建模的最新进展,利用最大熵原理从不同的数据中构建动态集成,同时解决不确定性和偏差。这些方法有助于解决异质性和解释低分辨率数据。最后,我们探讨了综合建模、增强采样和人工智能驱动的工具如何为缓慢、大规模的构象变化提供新的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current opinion in structural biology
生物-生化与分子生物学
CiteScore
12.20
自引率
2.90%
发文量
179
审稿时长
6-12 weeks
期刊介绍:
Current Opinion in Structural Biology (COSB) aims to stimulate scientifically grounded, interdisciplinary, multi-scale debate and exchange of ideas. It contains polished, concise and timely reviews and opinions, with particular emphasis on those articles published in the past two years. In addition to describing recent trends, the authors are encouraged to give their subjective opinion of the topics discussed.
In COSB, we help the reader by providing in a systematic manner:
1. The views of experts on current advances in their field in a clear and readable form.
2. Evaluations of the most interesting papers, annotated by experts, from the great wealth of original publications.
[...]
The subject of Structural Biology is divided into twelve themed sections, each of which is reviewed once a year. Each issue contains two sections, and the amount of space devoted to each section is related to its importance.
-Folding and Binding-
Nucleic acids and their protein complexes-
Macromolecular Machines-
Theory and Simulation-
Sequences and Topology-
New constructs and expression of proteins-
Membranes-
Engineering and Design-
Carbohydrate-protein interactions and glycosylation-
Biophysical and molecular biological methods-
Multi-protein assemblies in signalling-
Catalysis and Regulation
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


