Reinforced physiology-informed learning for image completion from partial-frame dynamic PET imaging
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
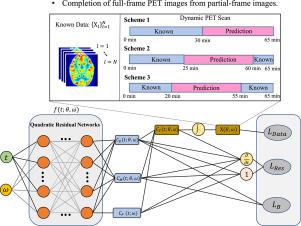
Dynamic positron emission tomography(PET) imaging using F-FDG typically requires over an hour to acquire a complete time series of images. Therefore, reducing dynamic PET scan time is crucial for minimizing errors caused by patient movement and increasing the throughput of the imaging equipment. However, shortening the scanning time will lead to the loss of images in some frames, affecting the accuracy of PET parameter estimation. In this paper, we proposed a method that combined physiology-informed learning with time-implicit neural representations for kinetic modeling and missing-frame dynamic PET image completion. Based on the two-tissue compartment model, three types of constraint terms were constructed for network training, including data terms, boundary terms, and reinforced physiology residual terms. The method works effectively without the need for specific training datasets, making it feasible even with limited data. Three commonly used scanning schemes were defined to verify the feasibility of the proposed method and the performance was evaluated based on simulation data and real rat data. The best-performing scheme was selected for detailed analysis of PET images and parameter maps on datasets of four human organs obtained from Biograph Vision Quadra. Our method outperforms traditional nonlinear least squares (NLLS) fitting in both reconstruction quality and computational efficiency. The metrics calculated from different organs, such as the brain (SSIM 0.98) and the thorax (PSNR 40), show that the proposed network can achieve promising performance.
从部分帧动态PET成像增强生理信息学习图像补全
使用18F-FDG的动态正电子发射断层扫描(PET)成像通常需要一个多小时才能获得完整的时间序列图像。因此,减少动态PET扫描时间对于最大限度地减少患者运动引起的错误和提高成像设备的吞吐量至关重要。然而,缩短扫描时间会导致某些帧的图像丢失,影响PET参数估计的准确性。在本文中,我们提出了一种将生理信息学习与时间隐式神经表征相结合的方法,用于动力学建模和缺帧动态PET图像补全。基于两组织隔室模型,构建了数据项、边界项和增强生理残差项三种约束项用于网络训练。该方法不需要特定的训练数据集就能有效地工作,即使在有限的数据下也是可行的。定义了三种常用的扫描方案,验证了所提方法的可行性,并基于仿真数据和真实大鼠数据对所提方法的性能进行了评价。选择性能最好的方案,对Biograph Vision Quadra获得的四个人体器官数据集的PET图像和参数图进行详细分析。该方法在重建质量和计算效率方面都优于传统的非线性最小二乘拟合。从不同器官,如大脑(SSIM > 0.98)和胸腔(PSNR > 40)计算的指标表明,所提出的网络可以取得良好的性能。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


