IterMask3D: Unsupervised anomaly detection and segmentation with test-time iterative mask refinement in 3D brain MRI
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Unsupervised anomaly detection and segmentation methods train a model to learn the training distribution as ‘normal’. In the testing phase, they identify patterns that deviate from this normal distribution as ‘anomalies’. To learn the ‘normal’ distribution, prevailing methods corrupt the images and train a model to reconstruct them. During testing, the model attempts to reconstruct corrupted inputs based on the learned ‘normal’ distribution. Deviations from this distribution lead to high reconstruction errors, which indicate potential anomalies. However, corrupting an input image inevitably causes information loss even in normal regions, leading to suboptimal reconstruction and an increased risk of false positives. To alleviate this, we propose , an iterative spatial mask-refining strategy designed for 3D brain MRI. We iteratively spatially mask areas of the image as corruption and reconstruct them, then shrink the mask based on reconstruction error. This process iteratively unmasks ‘normal’ areas to the model, whose information further guides reconstruction of ‘normal’ patterns under the mask to be reconstructed accurately, reducing false positives. In addition, to achieve better reconstruction performance, we also propose using high-frequency image content as additional structural information to guide the reconstruction of the masked area. Extensive experiments on the detection of both synthetic and real-world imaging artifacts, as well as segmentation of various pathological lesions across multiple MRI sequences, consistently demonstrate the effectiveness of our proposed method. Code is available at https://github.com/ZiyunLiang/IterMask3D.
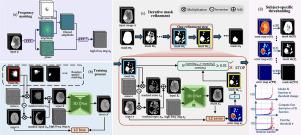
IterMask3D:基于测试时间迭代掩模细化的三维脑MRI无监督异常检测与分割
无监督异常检测和分割方法训练模型学习训练分布为“正态”。在测试阶段,他们将偏离正态分布的模式识别为“异常”。为了学习“正态”分布,流行的方法是破坏图像并训练一个模型来重建它们。在测试过程中,模型试图根据学习到的“正态”分布重建损坏的输入。偏离这一分布会导致较高的重建误差,这预示着潜在的异常。然而,即使在正常区域,损坏输入图像也不可避免地会导致信息丢失,从而导致次优重建和误报风险增加。为了缓解这一问题,我们提出了IterMask3D,这是一种针对3D脑MRI设计的迭代空间掩码细化策略。我们迭代地对图像的损坏区域进行空间掩码并进行重建,然后根据重建误差对掩码进行缩小。该过程迭代地揭开模型的“正常”区域,其信息进一步指导掩模下的“正常”模式的重建,以准确地重建,减少误报。此外,为了获得更好的重建性能,我们还提出使用高频图像内容作为附加的结构信息来指导被遮挡区域的重建。在检测合成和真实成像伪影以及在多个MRI序列中分割各种病理病变方面的大量实验一致证明了我们提出的方法的有效性。代码可从https://github.com/ZiyunLiang/IterMask3D获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


