Integration of transcriptome and targeted metabolome reveals regulatory and structural genes involved in anthocyanin biosynthesis of four Ceiba speciosa germplasms
IF 2.4
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
Ceiba speciosa has become a common landscape tree species in southern China due to its abundant petal colors and ornamental application values. Until now, there is little knowledge relevant to the molecular mechanisms involved in petal color formation and phenotypical variations in this species. In this study, transcriptome and targeted metabolome were integrated for four types of germplasm resources with different petal colors to identify candidate regulators, structural genes, and anthocyanins that might be involved in the formation of petal colors. Through qualitative and quantitative analyses of anthocyanins, the contents of two cyanidins (Cyanidin-3,5-O-diglucoside and Cyanidin-3-O-glucoside) were both consistent with color trends among four types of cultivars. By using RNA-seq, several structural genes, including six PALs, three F3Hs, one CHI, one 4CL, two CHSs, two DFRs, one ANS, and seven GT1s, were identified to be involved in the pathway of anthocyanin biosynthesis and were differentially expressed and highly correlated with the content of at least one compound. Then, 12 core transcription factors, including seven MYBs (MYB-33, MYB-37, MYB-80, MYB-92, MYB-111, MYB-112, and MYB-121), two C3Hs (C3H-105 and C3H-106), one EIL (EIL-10), one GRAS (GRAS-53), and one G2-like (G2-like-1), were obtained and possibly related to petal color differentiation together with other co-expressed transcription factors. In addition, the expression of numerous members belongs to an MYB-bHLH-WD40 module, and the main transcription factors involved in regulating anthocyanin biosynthesis were also significantly correlated with the content of five important anthocyanins, which have the highest average contents among all the samples. Molecular co-expression networks were constructed to display the relationship among TFs, structural genes, and metabolites. Our study lays the foundation for revealing the molecular mechanism involved in flower color production and variations in C. speciosa.
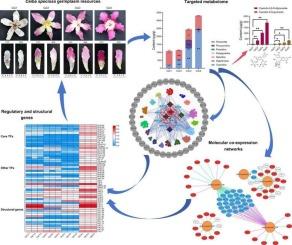
整合转录组和靶向代谢组揭示了四种Ceiba speciosa种质资源花青素合成的调控和结构基因
Ceiba speciosa因其丰富的花瓣颜色和观赏应用价值而成为中国南方常见的景观树种。到目前为止,有关该物种花瓣颜色形成和表型变异的分子机制的知识很少。本研究对4种不同花瓣颜色的种质资源进行转录组和靶向代谢组整合,鉴定可能参与花瓣颜色形成的候选调控因子、结构基因和花青素。通过花色苷的定性和定量分析,4种品种花色苷(花青素-3,5- o -二葡萄糖苷和花青素-3- o -葡萄糖苷)的含量均与花色趋势一致。通过RNA-seq,鉴定出6个PALs、3个F3Hs、1个CHI、1个4CL、2个CHSs、2个DFRs、1个ANS和7个GT1s等结构基因参与花青素生物合成途径,并与至少一种化合物的含量存在差异表达和高度相关。然后获得12个核心转录因子,包括7个myb (MYB-33、MYB-37、MYB-80、MYB-92、MYB-111、MYB-112和MYB-121), 2个c3h (C3H-105和C3H-106), 1个EIL (il -10), 1个GRAS (grass -53)和1个G2-like (G2-like-1),可能与其他共表达转录因子一起参与花瓣颜色分化。此外,许多成员的表达属于MYB-bHLH-WD40模块,参与调节花青素生物合成的主要转录因子也与5种重要花青素的含量显著相关,其中5种花青素的平均含量在所有样品中最高。构建分子共表达网络来显示tf、结构基因和代谢物之间的关系。本研究为揭示金盏花颜色产生和变异的分子机制奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Gene
生物-遗传学
CiteScore
6.10
自引率
2.90%
发文量
718
审稿时长
42 days
期刊介绍:
Gene publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


