Molecular Insights Into the Structure, Function, and Stability of the DNA Polymerase Processivity Factor From Mycobacterium tuberculosis
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
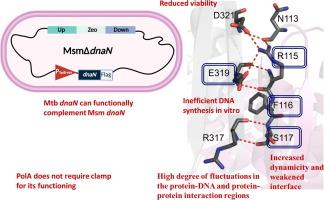
Emergence of drug resistance in Mycobacterium tuberculosis (Mtb) calls for newer drugs and drug targets. Essential proteins such as DNA polymerase (DNAP) processivity factor, also called sliding clamp (DnaN), are indispensable for bacterial survival, and are excellent drug targets. Here, we constructed a dnaN-conditional knockout in Mycobacterium smegmatis (MsmΔdnaN) and were able to successfully complement it with Mtb DnaN (DnaNMtb). To explore its structure–function–stability relationship, we generated Ala-substituted mutants of the DnaNMtb subunit-subunit interface, and identified R115, F116, and E319 as crucial for MsmΔdnaN survival in our complementation assay. We used biophysical, biochemical, and in silico molecular dynamics simulation methods to decipher the importance of these residues. We show that mutants exist as dimers, with lesser stability than wildtype. Except F116A, the mutants are largely folded with their CD profiles similar to wildtype. We also assembled and purified Mtb Clamp Loader Complex and used it to assess DNAP processivity function of DnaNMtb. Our in vitro DNA synthesis data show that PolAMtb does not interact with DnaNMtb, whereas E. coli Pol-I Klenow fragment shows enhanced DNA synthesis in presence of DnaNMtb, which was abolished by Griselimycin, an antibiotic that inhibits clamp-DNAP interaction. Interestingly, DnaNMtb mutants that did not complement loss of DnaN in MsmΔdnaN also did not support enhanced DNA synthesis by Klenow, corroborating our in vivo observation. We suggest that the Mtb clamp subunit-subunit interface is crucial for maintaining structure–function–stability, and thus can be used for the targeted development of small molecule inhibitors and peptidomimetics as potent drugs against tuberculosis.
结核分枝杆菌DNA聚合酶加工因子的结构、功能和稳定性的分子见解。
结核分枝杆菌(Mtb)耐药性的出现要求开发新的药物和药物靶点。必不可少的蛋白质,如DNA聚合酶(DNAP)加工因子,也称为滑动钳(DnaN),是细菌生存所必需的,也是很好的药物靶点。在这里,我们在耻垢分枝杆菌(MsmΔdnaN)中构建了一个DnaN条件敲除,并能够成功地与Mtb DnaN (DnaNMtb)互补。为了探索其结构-功能-稳定性关系,我们生成了DnaNMtb亚基-亚基界面的ala取代突变体,并在我们的互补实验中确定了R115, F116和E319对MsmΔdnaN存活至关重要。我们使用生物物理、生化和硅分子动力学模拟方法来破译这些残基的重要性。我们发现突变体以二聚体的形式存在,稳定性低于野生型。除F116A外,突变体大部分折叠,其CD谱与野生型相似。我们还组装并纯化了Mtb Clamp Loader复合物,并用它来评价DnaNMtb的DNAP加工功能。我们的体外DNA合成数据显示,PolAMtb不与DnaNMtb相互作用,而大肠杆菌Pol-I Klenow片段在DnaNMtb存在下显示出增强的DNA合成,而DnaNMtb被抑制钳-DNA相互作用的抗生素Griselimycin所消除。有趣的是,DnaNMtb突变体不补充MsmΔdnaN中DnaN的缺失,也不支持Klenow增强的DNA合成,证实了我们的体内观察。我们认为Mtb箝位亚基-亚基界面对于维持结构-功能稳定性至关重要,因此可以用于靶向开发小分子抑制剂和拟肽制剂作为抗结核病的有效药物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


