CeRTS: certainty retrieval token search in large language model clinical information extraction
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
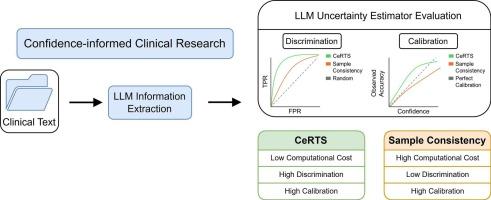
Large language models (LLMs) must effectively communicate their uncertainty to be viable in clinical settings. As such, the need for reliable uncertainty estimation grows increasingly urgent with the expanding use of LLMs for information extraction from electronic health records. Previous token-level uncertainty estimators have only used token probabilities within a single output sequence. Here, by leveraging the constraints of JSON output structure, we instead consider all likely sequences and their respective probabilities to obtain a more robust measure of model confidence. We develop Certainty Retrieval Token Search (CeRTS), a new uncertainty estimator for structured information extraction.
Methods
We evaluated CeRTS against a previous gold-standard uncertainty estimator when extracting clinical features from lung cancer discharge summaries across eight open-source LLMs. Calibration (Brier score) and discrimination (AUROC) were used to quantify performance.
Results
CeRTS surpassed the previous gold-standard estimator in discriminatory power across every model and achieved better calibration in most cases. CeRTS had the strongest agreement between model confidence and accuracy with Qwen-2.5.
Conclusion
CeRTS enhances LLM-based information extraction from unstructured clinical text by assigning well-calibrated confidence scores to each extracted item, providing medical researchers with a quantitative measure of reliability at minimal additional cost. Although its performance was generally robust, CeRTS struggled with DeepSeek-R1, which we attribute to the model’s Chain-of-Thought reasoning steps. Our evaluation focused on clinical data, but CeRTS can be applied to any domain requiring reliable uncertainty estimation.
CeRTS:确定性检索令牌搜索在大语言模型临床信息提取中的应用。
目的:大型语言模型(LLMs)必须有效地传达其不确定性,以便在临床环境中可行。因此,随着法学模型在电子健康记录信息提取中的广泛使用,对可靠的不确定性估计的需求日益迫切。以前的标记级不确定性估计器仅在单个输出序列中使用标记概率。在这里,通过利用JSON输出结构的约束,我们转而考虑所有可能的序列及其各自的概率,以获得更健壮的模型置信度度量。提出了确定性检索令牌搜索(CeRTS),一种新的结构化信息提取的不确定性估计方法。方法:在从八个开源llm的肺癌出院摘要中提取临床特征时,我们将CeRTS与先前的金标准不确定性估计器进行了评估。校正(Brier评分)和鉴别(AUROC)用于量化绩效。结果:CeRTS在每个模型的区分能力上都超过了以前的金标准估计器,并且在大多数情况下实现了更好的校准。CeRTS模型置信度和准确度与Qwen-2.5的一致性最强。结论:CeRTS增强了基于llm的非结构化临床文本信息提取,为每个提取项目分配了校准良好的置信度评分,以最小的额外成本为医学研究人员提供了定量的可靠性测量。尽管CeRTS的性能总体上是稳健的,但它在DeepSeek-R1上表现不佳,我们将其归因于该模型的思维链推理步骤。我们的评估侧重于临床数据,但CeRTS可以应用于任何需要可靠的不确定性估计的领域。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


