The C-terminal Domain of SARS-CoV-2 nsp8 is a Molten Globule in the Absence of Binding Partners
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The coronavirus genome is transcribed by a replication-transcription complex (RTC) containing the RNA polymerase plus additional cofactors. The cofactor nsp8 is an important component of the RTC in both alpha and betacoronaviruses required for nsp12 polymerase activity, complex stability, and recruitment of other RTC cofactors. Here we use NMR and other biophysical methods to characterize the structural features and oligomeric state of full-length nsp8 in solution. We show that the C-terminal domain of nsp8 has molten-globule like intrinsic disorder, while the N-terminal domain retains its folded structure in the absence of binding partners. Our data also shows a concentration-dependent association of nsp8 into dimers and possibly tetramers, but not larger molecular weight species. Upon binding nsp7, the C-terminal domain of nsp8 folds into a well-defined conformation consistent with available structures of the complex, while the linker region connecting the N- and C-terminal domains remains disordered.
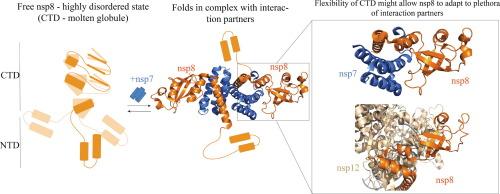
SARS-CoV-2 nsp8的c端结构域在没有结合伙伴的情况下呈熔融球状。
冠状病毒基因组由包含RNA聚合酶和其他辅助因子的复制转录复合体(RTC)转录。辅助因子nsp8是α和β冠状病毒中RTC的重要组成部分,对nsp12聚合酶活性、复合物稳定性和其他RTC辅助因子的募集都是必需的。本文采用核磁共振和其他生物物理方法表征了溶液中全长nsp8的结构特征和低聚态。我们发现nsp8的c端结构域具有熔融球样的内在无序,而n端结构域在没有结合伙伴的情况下保持其折叠结构。我们的数据还显示了nsp8与二聚体和可能的四聚体的浓度依赖性关联,但不是大分子量的物种。结合nsp7后,nsp8的c端结构域折叠成与复合物可用结构一致的明确构象,而连接N-和c端结构域的连接子区域仍然是无序的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


