Exploring foundation models for multi-class muscle segmentation in MR images of neuromuscular disorders: A comparative analysis of accuracy and uncertainty
IF 4.8
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background and Objective:
Deep learning (DL) models have shown promise for skeletal muscle (SM) segmentation in MR images, which is crucial for extracting biomarkers in neuromuscular disorders (NMDs). However, to ensure safe clinical use, models should provide uncertainty estimates, allowing radiologists to assess predictions and intervene when needed. Foundation Models (FMs) have the potential to play a significant role due to their strong generalization capabilities and well-calibrated predictions. However, their applicability in this context has not yet been explored. This study aims to develop an accurate and trustworthy technique by fine-tuning FMs to delineate fatty-infiltrated SM fascicles in NMD patients.
Methods:
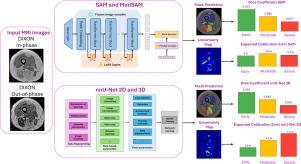
We fine-tuned Segment Anything Model (SAM) and MedSAM using two configurations – encoder/decoder and decoder only – and compared their performance against state-of-the-art 2D and 3D nnU-Net using a dataset of thigh MR images from 76 NMD patients, categorized into Early, Moderate, and Severe fatty infiltration groups. Accuracy was evaluated using the Dice Similarity Coefficient (DSC), while Uncertainty Quantification (UQ) was evaluated using the Expected Calibration Error (ECE) and the Negative Log-Likelihood (NLL). Deep Ensembles were used to convey epistemic uncertainty in addition to the aleatoric counterpart.
Results:
SAM’s fine-tuned encoder/decoder outperformed nnU-Net 3D in Moderate and Severe cases (DSC: 0.886 vs 0.883 and 0.857 vs 0.850) and was comparable in Early (DSC: 0.925). MedSAM did not show an advantage over SAM. Regarding UQ, SAM exhibited superior calibration in Moderate and Severe groups (ECE: 3.6% vs. 5.1% and 3.3% vs. 7.1%),
Conclusions:
In conclusion, our findings demonstrate that fine-tuning SAM yields superior performance, considering both accuracy and UQ metrics, highlighting its enhanced reliability in challenging NMD imaging scenarios.
探索神经肌肉疾病MR图像中多类别肌肉分割的基础模型:准确性和不确定性的比较分析
背景与目的:深度学习(DL)模型在MR图像的骨骼肌(SM)分割中显示出前景,这对于提取神经肌肉疾病(nmd)的生物标志物至关重要。然而,为了确保安全的临床使用,模型应该提供不确定性估计,允许放射科医生评估预测并在需要时进行干预。基础模型(FMs)由于其强大的泛化能力和良好的校准预测而具有发挥重要作用的潜力。然而,它们在这方面的适用性尚未得到探讨。本研究旨在开发一种准确可靠的技术,通过微调FMs来描绘NMD患者脂肪浸润的SM束。方法:我们使用编码器/解码器和仅解码器两种配置对片段任意模型(SAM)和MedSAM进行微调,并使用76名NMD患者的大腿MR图像数据集将其性能与最先进的2D和3D nnU-Net进行比较,这些患者被分为早期、中度和重度脂肪浸润组。使用骰子相似系数(DSC)评估准确性,而使用预期校准误差(ECE)和负对数似然(NLL)评估不确定性量化(UQ)。除了任意对应物外,还使用深度集成来传达认知不确定性。结果:SAM的微调编码器/解码器在中度和重度患者中的表现优于nnU-Net 3D (DSC: 0.886 vs 0.883和0.857 vs 0.850),在早期患者中表现相当(DSC: 0.925)。MedSAM没有表现出优于SAM的优势。关于UQ, SAM在中度和重度组表现出更好的校准(ECE: 3.6% vs. 5.1%, 3.3% vs. 7.1%)。结论:总之,我们的研究结果表明,考虑到准确性和UQ指标,微调SAM产生了更好的性能,突出了其在具有挑战性的NMD成像场景中的增强可靠性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


