Improving large language models for adverse drug reactions named entity recognition via error correction prompt engineering
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
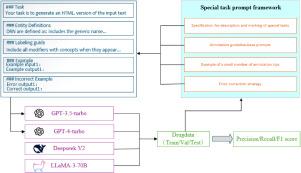
The monitoring and analysis of adverse drug reactions (ADRs ) are important for ensuring patient safety and improving treatment outcomes. Accurate identification of drug names, drug components, and ADR entities during named entity recognition (NER) processes is essential for ensuring drug safety and advancing the integration of drug information. Given that existing medical name entity recognition technologies rely on large amounts of manually annotated data for training, they are often less effective when applied to adverse drug reactions due to significant data variability and the high similarity between drug names. This paper proposes a prompt template for ADR that integrates error correction examples. The prompt template includes: 1. Basic prompts with task descriptions, 2. Annotated entity explanations, 3. Annotation guidelines, 4. Annotated samples for few-shot learning, 5. Error correction examples. Additionally, it integrates complex ADR data from the web and constructs a corpus containing three types of entities (drug name, drug components, and adverse drug reactions) using the Begin, Inside, Outside (BIO) annotation method. Finally, we evaluate the effectiveness of each prompt and compare it with the fine-tuned Large Language Model Meta AI (LLaMA) model and the DeepSeek model. Experimental results show that under this prompt template, the F1 score of GPT-3.5 increased from 0.648 to 0.887, and that of GPT-4 increased from 0.757 to 0.921. It is significantly better than the fine-tuned LLaMA model and DeepSeek model. It demonstrates the superiority of the proposed method, and provides a solid foundation for extracting drug-related entity relationships and building knowledge graphs.
通过纠错提示工程改进药物不良反应命名实体识别的大型语言模型
药物不良反应(adr)的监测和分析对于确保患者安全和改善治疗效果非常重要。在命名实体识别(NER)过程中,准确识别药品名称、药物成分和ADR实体对于确保药品安全和推进药品信息整合至关重要。鉴于现有的医学名称实体识别技术依赖于大量人工标注的数据进行训练,由于数据的显著可变性和药品名称之间的高度相似性,它们在应用于药物不良反应时往往效果较差。本文提出了一个集成纠错实例的ADR提示模板。提示符模板包括:1。2、基本提示和任务描述。注释实体解释;注释指南;5.少射学习的标注样本;纠错示例。此外,它集成了来自网络的复杂ADR数据,并使用Begin, Inside, Outside (BIO)注释方法构建了一个包含三种类型实体(药物名称,药物成分和药物不良反应)的语料库。最后,我们评估了每个提示的有效性,并将其与经过微调的大型语言模型元AI (LLaMA)模型和DeepSeek模型进行了比较。实验结果表明,在该提示模板下,GPT-3.5的F1评分从0.648提高到0.887,GPT-4的F1评分从0.757提高到0.921。它明显优于经过微调的LLaMA模型和DeepSeek模型。验证了该方法的优越性,为药物相关实体关系的提取和知识图谱的构建提供了坚实的基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


