Is the vaccination-induced B cell receptor repertoire predictable?
引用次数: 0
Abstract
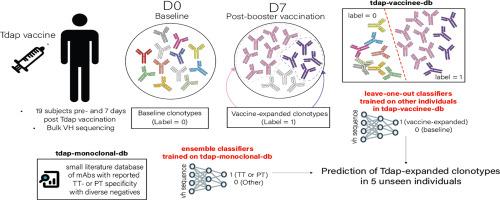
Vaccines trigger an immune response that results in a population of memory cells that can quickly respond to subsequent antigen re-encounters. Most vaccines are designed to induce memory B cells with vaccine-specific B cell receptors (BCRs). Post-vaccination, clonal expansion of B cells results in measurably expanded vaccine-specific BCR clonotypes. We set out to determine to what extent it is predictable which specific BCR clonotypes are vaccine-induced in an individual. We sequenced the BCR heavy chain repertoire in a cohort of 19 individuals prior- and 7 days post Tdap booster vaccination. We tested two modalities to predict which clonotypes were expanded post-vaccination: first, we utilized a small database of monoclonal antibodies with known specificity to Tdap vaccine antigens and tested various sequence look-up methods, identifying clonal look-up as the best method. We then utilized a leave-one-out approach in which expanded clonotypes in one individual were predicted using data from other members of the cohort. The second approach significantly outperformed the first, indicating that BCR clonotype expansion can be learned across subjects. These results support the utility of systematically collecting BCR specificity data through efforts like the Immune Epitope database and highlight the limitations on general prediction approaches resulting from relatively small dataset sizes for BCRs with known specificities. Additionally, our study provides 1) a comparison of several BCR specificity prediction methods, 2) a dataset that can be used for benchmarking of subsequent methods, and 3) a methodological framework for comparing BCR repertoires pre- and post-vaccination.
疫苗诱导的B细胞受体库可预测吗?
疫苗会引发免疫反应,导致记忆细胞群迅速对随后再次遇到的抗原做出反应。大多数疫苗被设计成用疫苗特异性B细胞受体(BCRs)诱导记忆B细胞。接种后,B细胞克隆扩增导致可测量扩增的疫苗特异性BCR克隆型。我们着手确定在多大程度上可以预测疫苗在个体中诱导的特定BCR克隆型。我们对接种Tdap加强疫苗前和接种后7天的19名个体的BCR重链库进行了测序。我们测试了两种方法来预测疫苗接种后扩增的克隆型:首先,我们利用了一个已知Tdap疫苗抗原特异性的小型单克隆抗体数据库,并测试了各种序列查找方法,确定克隆查找方法是最佳方法。然后,我们使用了一种“留一”方法,其中使用来自队列其他成员的数据预测一个个体的扩展克隆型。第二种方法明显优于第一种方法,表明BCR克隆型扩增可以跨受试者学习。这些结果支持通过免疫表位数据库等工作系统地收集BCR特异性数据的效用,并突出了由于已知特异性的BCR相对较小的数据集大小而导致的一般预测方法的局限性。此外,我们的研究提供了1)几种BCR特异性预测方法的比较,2)可用于对后续方法进行基准测试的数据集,以及3)比较接种前和接种后BCR库的方法框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Immunoinformatics (Amsterdam, Netherlands)
Immunology, Computer Science Applications
自引率
0.00%
发文量
0
审稿时长
60 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


