Source independent multiple-domain adaptation for knee osteoarthritis cartilage and meniscus segmentation in clinical magnetic resonance imaging
IF 6.9
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background
Generalized knee tissue segmentation, such as cartilage and meniscus in magnetic resonance imaging (MRI), plays a vital role in the clinical assessment of knee osteoarthritis (OA). However, domain variability between MRI datasets poses a significant challenge for the application of robust segmentation methods in real-world clinical settings. Existing unsupervised domain adaptation (UDA) approaches, which rely on one-to-one assumptions between the source and target domains, often fail to preserve knee tissues such as cartilage and meniscus, which are critical for OA diagnosis in diverse clinical settings.
Methods
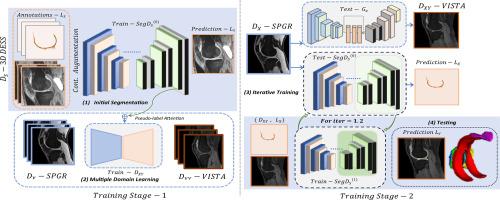
We propose a source-independent segmentation approach tailored for multi-domain knee MRI datasets. Our method emphasizes knee tissue regions to reduce domain gaps and label inconsistencies. By introducing a stepwise adaptation strategy, segmentation performance was refined progressively from intermediate domains to the final target domain. Pseudo-label attention mechanisms were integrated into the adaptation pipeline, enabling iterative fine-tuning of domain-specific segmentations while leveraging unidirectional generative adversarial networks to enhance tissue-specific adaptation. This iterative training process ensures the generation of reliable pseudo-labels, thereby improving segmentation accuracy in diverse clinical MRI datasets.
Results
We demonstrated the effectiveness of our approach on the OA initiative dataset as the source domain and self-collected, T1-weighted fast field echo (T1FFE) as the intermediate domain and three-dimensional fast spin echo (3D FSE) as the final target domain. Our method achieved an average dice scores of 0.8701 and 0.7990 for source and target domains, respectively, surpassing the typical UDA methods explored in our experiments.
Conclusion
The experiments conducted on clinical MRI data, spanning OA severity from healthy knees to KL Grades 1–4, validated the effectiveness of the proposed domain adaptation method in precise segmentation of the cartilage and meniscus.
临床磁共振成像中膝关节骨关节炎软骨和半月板分割的源独立多域自适应
广泛的膝关节组织分割,如磁共振成像(MRI)中的软骨和半月板,在膝关节骨关节炎(OA)的临床评估中起着至关重要的作用。然而,MRI数据集之间的区域可变性对现实世界临床环境中鲁棒分割方法的应用提出了重大挑战。现有的无监督域适应(UDA)方法依赖于源域和目标域之间的一对一假设,通常无法保护膝关节组织,如软骨和半月板,而这些组织在不同的临床环境中对OA诊断至关重要。方法针对多域膝关节MRI数据集,提出一种与源无关的分割方法。我们的方法强调膝关节组织区域,以减少区域间隙和标记不一致。通过引入逐步适应策略,从中间域到最终目标域的分割性能逐步得到改进。伪标签注意机制被集成到适应管道中,实现了特定领域分割的迭代微调,同时利用单向生成对抗网络来增强组织特异性适应。这种迭代训练过程保证了生成可靠的伪标签,从而提高了不同临床MRI数据集的分割精度。结果以OA主动数据集为源域,以自采集数据集为中间域,以t1加权快速场回波(T1FFE)为中间域,以三维快速自旋回波(3D FSE)为最终目标域,验证了该方法的有效性。我们的方法在源域和目标域的平均骰子得分分别为0.8701和0.7990,超过了我们在实验中探索的典型UDA方法。基于临床MRI数据进行的实验,涵盖了从健康膝关节到KL分级1-4级的OA严重程度,验证了所提出的区域适应方法在软骨和半月板精确分割方面的有效性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Intelligent medicine
Surgery, Radiology and Imaging, Artificial Intelligence, Biomedical Engineering
CiteScore
5.20
自引率
0.00%
发文量
19
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


