Knowledge graph-driven curation of heme-TLR4 interactions in inflammatory pathways
IF 3.2
2区 化学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Heme, a vital iron-containing molecule, serves fundamental roles in oxygen transport and electron transfer but also acts as an extracellular signaling entity, significantly influencing inflammatory responses. Elevated levels of labile heme resulting from hemolytic events or therapeutic treatments may activate inflammatory signaling pathways, particularly through the Toll-like receptor 4 (TLR4). In this study, we systematically expanded the previously developed Heme Knowledge Graph (HemeKG) to comprehensively incorporate recent findings regarding heme-TLR4 interactions. By employing rigorous literature curation and validation using Biological Expression Language (BEL) standards and the e:BEL Python package, we successfully integrated newly identified molecular entities, notably activator protein 1 (AP-1), interleukin-12 (IL-12), cluster of differentiation 80 (CD80), cluster of differentiation 86 (CD86), and chemokine (C-X-C motif) ligand 1 (CXCL1), into the existing HemeKG framework. Pathway enrichment analysis across Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, and WikiPathways databases robustly supported these integrations, consistently identifying significant enrichment of the TLR4 signaling cascade. The updated HemeKG thus provides an integrated and predictive platform, enhancing our understanding of the complex interactions between heme-driven inflammatory pathways and metabolic dysregulation.
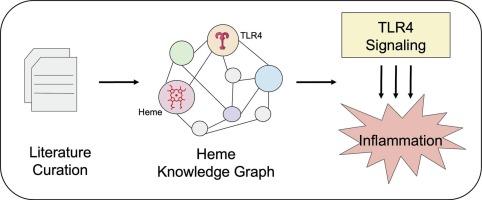
炎性通路中血红素- tlr4相互作用的知识图谱驱动管理
血红素是一种重要的含铁分子,在氧转运和电子转移中起着重要作用,但也作为细胞外信号实体,显著影响炎症反应。溶血事件或治疗引起的不稳定血红素水平升高可能激活炎症信号通路,特别是通过toll样受体4 (TLR4)。在这项研究中,我们系统地扩展了先前开发的血红素知识图谱(HemeKG),以全面纳入有关血红素- tlr4相互作用的最新发现。通过使用生物表达语言(BEL)标准和e:BEL Python软件包进行严格的文献整理和验证,我们成功地将新发现的分子实体,特别是激活蛋白1 (AP-1)、白细胞介素-12 (IL-12)、分化簇80 (CD80)、分化簇86 (CD86)和趋化因子(C-X-C基序)配体1 (CXCL1)整合到现有的HemeKG框架中。京都基因与基因组百科全书(KEGG)、Reactome和WikiPathways数据库的通路富集分析有力地支持了这些整合,一致地确定了TLR4信号级联的显著富集。因此,更新后的HemeKG提供了一个集成的预测平台,增强了我们对血红素驱动的炎症途径与代谢失调之间复杂相互作用的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Journal of Inorganic Biochemistry
生物-生化与分子生物学
CiteScore
7.00
自引率
10.30%
发文量
336
审稿时长
41 days
期刊介绍:
The Journal of Inorganic Biochemistry is an established international forum for research in all aspects of Biological Inorganic Chemistry. Original papers of a high scientific level are published in the form of Articles (full length papers), Short Communications, Focused Reviews and Bioinorganic Methods. Topics include: the chemistry, structure and function of metalloenzymes; the interaction of inorganic ions and molecules with proteins and nucleic acids; the synthesis and properties of coordination complexes of biological interest including both structural and functional model systems; the function of metal- containing systems in the regulation of gene expression; the role of metals in medicine; the application of spectroscopic methods to determine the structure of metallobiomolecules; the preparation and characterization of metal-based biomaterials; and related systems. The emphasis of the Journal is on the structure and mechanism of action of metallobiomolecules.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


