Rigorous integration of single-cell ATAC-seq data using regularized barycentric mapping
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
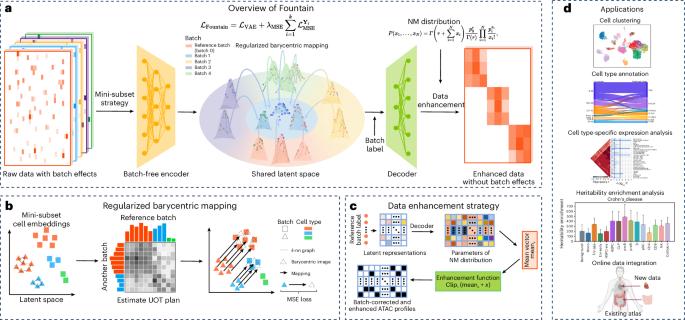
Single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) deciphers genome-wide chromatin accessibility, providing profound insights into gene regulation mechanisms. With the rapid advance of sequencing technologies, scATAC-seq data typically encompass numerous samples from various conditions, resulting in complex batch effects, thus necessitating reliable integration tools. While numerous batch integration tools exist for single-cell RNA sequencing data, inherent data characteristic differences limit their effectiveness on scATAC-seq data. Existing integration methods for scATAC-seq data suffer from several fundamental limitations, such as disrupting the biological heterogeneity and focusing solely on low-dimensional correction, which may distort data and hinder downstream analysis. Here we propose Fountain, a deep learning framework for scATAC-seq data integration via rigorous barycentric mapping. Barycentric mapping transforms one data distribution to another in a principled and effective manner through optimal transport. By regularizing barycentric mapping with geometric data information, Fountain achieves accurate batch alignment while preserving biological heterogeneity. Comprehensive experiments across diverse real-world datasets demonstrate the advantages of Fountain over existing methods in batch correction and biological conservation. In addition, the trained Fountain model can integrate data from new batches alongside already integrated data without retraining, enabling continuous online data integration. Moreover, Fountain’s reconstruction strategy generates batch-corrected ATAC profiles, improving the capture of cellular heterogeneity and revealing cell-type-specific implications such as expression enrichment analysis and partitioned heritability analysis. Zhu, Hua and Chen propose Fountain, a deep learning framework for batch integration of scATAC-seq data that utilizes regularized barycentric mapping. It preserves biological heterogeneity, enabling online and original dimensionality integration.
严格整合单细胞ATAC-seq数据使用正则化质心映射
利用测序技术(scATAC-seq)对转座酶可及染色质进行单细胞分析,可以破译全基因组染色质可及性,为基因调控机制提供深刻的见解。随着测序技术的快速发展,scATAC-seq数据通常包含来自不同条件的大量样本,导致复杂的批处理效果,因此需要可靠的集成工具。虽然存在许多用于单细胞RNA测序数据的批量集成工具,但固有的数据特征差异限制了它们在scATAC-seq数据上的有效性。现有的scATAC-seq数据整合方法存在一些根本性的局限性,例如破坏了生物异质性,只关注低维校正,这可能会扭曲数据并阻碍下游分析。在这里,我们提出了Fountain,这是一个通过严格的重心映射进行scATAC-seq数据集成的深度学习框架。重心映射通过优化传输,以原则和有效的方式将一个数据分布转换为另一个数据分布。通过正则化几何数据信息的质心映射,Fountain在保持生物异质性的同时实现了精确的批量对齐。在不同的真实世界数据集上进行的综合实验表明,Fountain在批量校正和生物保护方面优于现有方法。此外,经过训练的Fountain模型可以将新批次的数据与已经集成的数据集成在一起,而无需重新训练,从而实现持续的在线数据集成。此外,Fountain的重建策略生成了批量校正的ATAC谱,改善了细胞异质性的捕获,揭示了细胞类型特异性的含义,如表达富集分析和分区遗传力分析。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


