Acquisition-independent deep learning for quantitative MRI parameter estimation using neural controlled differential equations
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Deep learning has proven to be a suitable alternative to least squares (LSQ) fitting for parameter estimation in various quantitative MRI (QMRI) models. However, current deep learning implementations are not robust to changes in MR acquisition protocols. In practice, QMRI acquisition protocols differ substantially between different studies and clinical settings. The lack of generalizability and adoptability of current deep learning approaches for QMRI parameter estimation impedes the implementation of these algorithms in clinical trials and clinical practice. Neural Controlled Differential Equations (NCDEs) allow for the sampling of incomplete and irregularly sampled data with variable length, making them ideal for use in QMRI parameter estimation. In this study, we show that NCDEs can function as a generic tool for the accurate estimation of QMRI parameters, regardless of QMRI sequence length, configuration of independent variables and QMRI forward model (variable flip angle T1-mapping, intravoxel incoherent motion MRI, dynamic contrast-enhanced MRI). NCDEs achieved lower mean squared error than LSQ fitting in low-SNR simulations and in vivo in challenging anatomical regions like the abdomen and leg, but this improvement was no longer evident at high SNR. When NCDEs improve parameter estimation, they tend to do so by reducing the variance in estimation errors. These findings suggest that NCDEs offer a robust approach for reliable QMRI parameter estimation, especially in scenarios with high uncertainty or low image quality. We believe that with NCDEs, we have solved one of the main challenges for using deep learning for QMRI parameter estimation in a broader clinical and research setting.
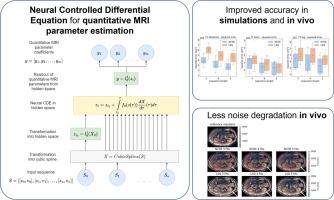
利用神经控制微分方程进行定量MRI参数估计的非获取深度学习
深度学习已被证明是各种定量MRI (QMRI)模型参数估计的最小二乘(LSQ)拟合的合适替代方法。然而,目前的深度学习实现对MR获取协议的变化并不健壮。在实践中,QMRI采集方案在不同的研究和临床环境中存在很大差异。目前用于QMRI参数估计的深度学习方法缺乏通用性和可接受性,阻碍了这些算法在临床试验和临床实践中的实施。神经控制微分方程(NCDEs)允许对可变长度的不完整和不规则采样数据进行采样,使其非常适合用于QMRI参数估计。在这项研究中,我们发现NCDEs可以作为一种通用工具,用于准确估计QMRI参数,而不受QMRI序列长度、自变量配置和QMRI正演模型(可变翻转角t1映射、体素内非相干运动MRI、动态对比度增强MRI)的影响。NCDEs在低信噪比模拟和腹部和腿部等具有挑战性的解剖区域的体内拟合中均方误差低于LSQ拟合,但在高信噪比下这种改善不再明显。当ncde改进参数估计时,它们倾向于通过减少估计误差的方差来做到这一点。这些发现表明,NCDEs为可靠的QMRI参数估计提供了一种鲁棒方法,特别是在高不确定性或低图像质量的情况下。我们相信,通过NCDEs,我们已经解决了在更广泛的临床和研究环境中使用深度学习进行QMRI参数估计的主要挑战之一。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


