Proteoformics: Current status and future perspectives
IF 2.8
2区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
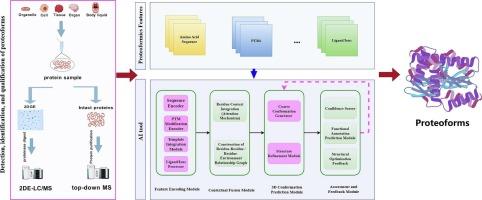
Proteoforms represent the ultimate structural/functional forms of a gene product, defined by multiple factors, including amino acid sequences, post-translational modifications, spatial conformations, and interactions with other molecules. The human proteoform diversity significantly exceeds the number of human genes/transcripts, emphasizing the need for advanced analytical methods to characterize this complexity. Two-dimensional gel electrophoresis-liquid chromatography/mass spectrometry (2DE-LC/MS) and top-down MS (TD-MS) are complementary to detect, identify, and quantify the large-scale proteoforms. The emerging AI tools for structural biology such as AlphaFold 3 and D-I-TASSER will enable proteoformics to be high-throughput and precisely predict spatial conformations and molecular interactions. Integrating the large-scale experimental data derived from 2DE-LC/MS and TD-MS with AI-driven high-throughput structural analysis paves the way to deeply understand proteoform diversity and functionality. The combination of advanced 2DE-LC/MS, TD-MS, and AI-driven structural analysis represents a pivotal advancement in proteoformics. This integrated approach enables the comprehensive profiling of proteoforms, providing critical insights into their roles in health care. Such advancements hold promise for predictive, preventive, and personalized medicine, particularly through biomarker discovery and therapeutic target identification. Future developments in high-throughput capabilities and dynamic modeling are expected to address current challenges and further expand the applicability of proteoformics in biological and clinical research.
Significance
Proteoformics is the future of proteomics, whose two main complementary analytical approaches are 2DE-LC/MS and TD-MS. The AI-driven large-cale structural analysis enables to high-throughput and precisely analyze spatial conformations and molecular interactions of proteoforms, which helps to deeply understand proteoform diversity and functionality. Proteoformics holds transformative potential to uncover biomarkers, guide targeted therapies, and advance predictive diagnosis in the context of personalized medicine.
蛋白质组学:现状与未来展望
蛋白质形态代表了基因产物的最终结构/功能形式,由多种因素决定,包括氨基酸序列、翻译后修饰、空间构象以及与其他分子的相互作用。人类蛋白质形态的多样性大大超过了人类基因/转录本的数量,强调需要先进的分析方法来表征这种复杂性。二维凝胶电泳-液相色谱/质谱法(2DE-LC/MS)和自上而下质谱法(TD-MS)在检测、鉴定和定量大规模蛋白质形态方面是互补的。新兴的结构生物学人工智能工具,如AlphaFold 3和D-I-TASSER,将使蛋白质形成学具有高通量,并精确预测空间构象和分子相互作用。将来自2DE-LC/MS和TD-MS的大规模实验数据与人工智能驱动的高通量结构分析相结合,为深入了解蛋白质形态多样性和功能铺平了道路。先进的2DE-LC/MS, TD-MS和ai驱动的结构分析的结合代表了蛋白质组学的关键进步。这种综合方法能够全面分析变形形态,为其在卫生保健中的作用提供关键见解。这些进步为预测性、预防性和个性化医学带来了希望,特别是通过生物标志物的发现和治疗靶点的识别。高通量能力和动态建模的未来发展有望解决当前的挑战,并进一步扩大蛋白质组学在生物学和临床研究中的适用性。蛋白质组学是蛋白质组学的未来,其两种主要互补的分析方法是2DE-LC/MS和TD-MS。人工智能驱动的大尺度结构分析能够高通量、精确地分析变形形态的空间构象和分子相互作用,有助于深入了解变形形态的多样性和功能。蛋白质组学在揭示生物标志物、指导靶向治疗和在个性化医疗背景下推进预测性诊断方面具有变革性潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of proteomics
生物-生化研究方法
CiteScore
7.10
自引率
3.00%
发文量
227
审稿时长
73 days
期刊介绍:
Journal of Proteomics is aimed at protein scientists and analytical chemists in the field of proteomics, biomarker discovery, protein analytics, plant proteomics, microbial and animal proteomics, human studies, tissue imaging by mass spectrometry, non-conventional and non-model organism proteomics, and protein bioinformatics. The journal welcomes papers in new and upcoming areas such as metabolomics, genomics, systems biology, toxicogenomics, pharmacoproteomics.
Journal of Proteomics unifies both fundamental scientists and clinicians, and includes translational research. Suggestions for reviews, webinars and thematic issues are welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


