Strand-specific quantification of 8-oxo-dG and apurinic sites via Ligation-Dependent Probe Amplification (LPA)
IF 11.9
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Oxidative DNA damage, characterized by the prominent lesion 8-oxo-7,8-dihydroguanine (8-oxo-dG), is linked to mutagenesis and genome instability. Accurately mapping these lesions with strand specificity and high resolution remains a major challenge, limiting our understanding of damage dynamics during transcription and repair. Here, we introduce a novel, highly sensitive ligation-dependent probe amplification (LPA) method that enables quantitative, strand-specific analysis of 8-oxo-dG and apurinic (AP) sites at single-nucleotide resolution. Our technique uses enzymatic digestion, highly selective ligation, and quantitative PCR to distinguish damaged from intact DNA strands, offering detailed insight into lesion localization and repair kinetics. When applied to estrogen-stimulated breast cancer cells, LPA reveals asymmetric, strand-specific guanine oxidation during transcriptional activation, characterized by rapid repair of the transcribed strand and more persistent damage on the non-transcribed strand. Our findings show that oxidative lesions are dynamically regulated by biological stimuli, reflecting a finely tuned balance between repair and damage buildup. This LPA approach is a powerful tool for exploring the complex relationship among redox signaling, DNA damage, and transcription regulation, furthering our understanding of redox-driven genome modulation in health and disease.
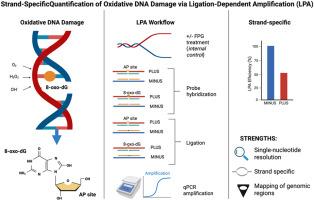
通过连接依赖探针扩增(LPA)对8-oxo-dG和apurinic位点进行链特异性定量
以8-氧-7,8-二氢鸟嘌呤(8-氧- dg)损伤为特征的氧化性DNA损伤与突变和基因组不稳定有关。具有链特异性和高分辨率的准确绘制这些病变仍然是一个主要挑战,限制了我们对转录和修复过程中损伤动力学的理解。在这里,我们介绍了一种新颖的,高度敏感的连接依赖探针扩增(LPA)方法,可以在单核苷酸分辨率下定量,链特异性分析8-oxo-dG和apurinic (AP)位点。我们的技术使用酶切、高选择性连接和定量PCR来区分受损的DNA链和完整的DNA链,提供详细的损伤定位和修复动力学。当应用于雌激素刺激的乳腺癌细胞时,LPA在转录激活过程中显示出不对称的、链特异性的鸟嘌呤氧化,其特征是转录链的快速修复和非转录链的更持久的损伤。我们的研究结果表明,氧化损伤受到生物刺激的动态调节,反映了修复和损伤积累之间的微妙平衡。这种LPA方法是探索氧化还原信号、DNA损伤和转录调控之间复杂关系的有力工具,进一步加深了我们对氧化还原驱动的基因组在健康和疾病中的调节的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Redox Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-
CiteScore
19.90
自引率
3.50%
发文量
318
审稿时长
25 days
期刊介绍:
Redox Biology is the official journal of the Society for Redox Biology and Medicine and the Society for Free Radical Research-Europe. It is also affiliated with the International Society for Free Radical Research (SFRRI). This journal serves as a platform for publishing pioneering research, innovative methods, and comprehensive review articles in the field of redox biology, encompassing both health and disease.
Redox Biology welcomes various forms of contributions, including research articles (short or full communications), methods, mini-reviews, and commentaries. Through its diverse range of published content, Redox Biology aims to foster advancements and insights in the understanding of redox biology and its implications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


