Biomedical knowledge graph verification with multitask learning architectures
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective
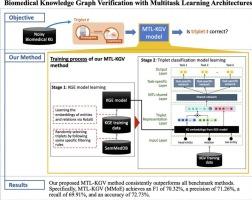
Large-scale biomedical KGs, typically constructed using automated entity-relation extraction methods from vast amounts of textual documents, often contain erroneous biomedical triplets, which raises concerns about their quality. Using such noisy KGs in downstream applications can compromise the validity of biomedical research and lead to inaccurate conclusions. This study aims to design an effective knowledge graph verification (KGV) method to determine the correctness of triplets in biomedical KGs, enabling the removal of erroneous triplets identified through the proposed method.
Methods
We propose a multitask-learning-based KGV (referred to as the MTL-KGV) method, which includes two key stages: 1) KG embedding (KGE) learning and (2) triplet classification model learning. In addition, we explore three types of multitask learning (MTL) architectures—hard parameter sharing (HPS), multi-gate mixture-of-experts (MMoE), and customized gate control (CGC)—for triplet classification model learning.
Results
Using SemMedDB as a data source to construct a large-scale KG for KGE training and a dataset of 6,427 biomedical triplets annotated by a domain expert, we empirically evaluate the effectiveness of our proposed MTL-KGV method by comparing it to several benchmark methods. Our evaluation results indicate that all three versions of our proposed MTL-KGV method consistently outperform the benchmark methods. Moreover, our proposed method with the MMoE multitask learning architecture emerges as the most effective for detecting erroneous biomedical triplets.
Conclusion
This work contributes to KGV research by introducing a multitask learning framework tailored for KGV. The proposed MTL-KGV method improves the quality of biomedical KGs, thereby supporting downstream applications and advancing biomedical research that relies on these biomedical KGs.
基于多任务学习架构的生物医学知识图谱验证
大型生物医学知识库通常使用自动化实体关系提取方法从大量文本文档中构建,通常包含错误的生物医学三元组,这引起了对其质量的担忧。在下游应用中使用这种噪声kg会损害生物医学研究的有效性,并导致不准确的结论。本研究旨在设计一种有效的知识图谱验证(knowledge graph verification, KGV)方法来确定生物医学知识图谱中三胞胎的正确性,从而去除通过该方法识别出的错误三胞胎。方法提出了一种基于多任务学习的KGV方法(简称MTL-KGV),该方法包括两个关键阶段:1)KG嵌入(KGE)学习和(2)三元组分类模型学习。此外,我们探索了三种类型的多任务学习(MTL)架构-硬参数共享(HPS),多门混合专家(MMoE)和定制门控制(CGC) -用于三重分类模型学习。结果以SemMedDB为数据源,构建了用于KGE训练的大规模KG和由领域专家注释的6,427个生物医学三联体数据集,通过与几种基准方法进行比较,实证评估了所提出的MTL-KGV方法的有效性。我们的评估结果表明,我们提出的MTL-KGV方法的所有三个版本都始终优于基准方法。此外,我们提出的基于MMoE多任务学习架构的方法对于检测错误的生物医学三胞胎是最有效的。本研究引入了一个针对KGV的多任务学习框架,为KGV的研究做出了贡献。提出的MTL-KGV方法提高了生物医学kg的质量,从而支持下游应用并推进依赖于这些生物医学kg的生物医学研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


