Sequence/structural/functional relationships between Ganoderma fungal immunomodulatory proteins (gFIPs) and proteins involved in the modulation of immune response
IF 2.7
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Fungal Immunomodulatory Proteins from Ganoderma species (gFIPs) have garnered significant interest due to their potential therapeutic applications in modulating immune responses. This study investigates the sequence, structural, and functional relationships of gFIPs with other proteins involved in immune modulation. Utilizing molecular modelling, multiple sequence alignments, and structural superimposition, we analysed two FIP crystallized structures (PDB IDs: 3F3H and 3KCW) alongside homologous sequences from various taxonomic groups. Our results reveal conserved motifs across fungal, bacterial, and human sequences, indicating potential functional similarities. Comparative structural analysis highlights significant conservation in FIP architecture, with variations primarily in the N-terminal regions. Notably, structural alignment with bacterial toxins, such as ADP-ribosylating binary toxin from Clostridium difficile or protective antigen of Anthrax toxin from Bacillus anthracis suggests mechanistic insights into FIP’s immunomodulatory actions. Structural similarities between gFIPs and immune-related proteins, such as bacterial toxin-binding domains, antibody fragments, T-cell receptor components, and immune checkpoint regulators (PD-1) suggest their potential involvement in immune response/inflammation signalling pathways. This comprehensive analysis elucidates the structural basis for the diverse biological activities of gFIPs and underscores their potential as therapeutic agents in immune-related diseases.
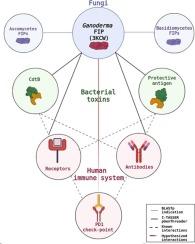
灵芝真菌免疫调节蛋白(gFIPs)与免疫应答调节蛋白之间的序列/结构/功能关系
来自灵芝物种的真菌免疫调节蛋白(gFIPs)由于其在调节免疫反应方面的潜在治疗应用而引起了极大的兴趣。本研究探讨了gFIPs与其他参与免疫调节的蛋白的序列、结构和功能关系。利用分子模型、多序列比对和结构叠加,我们分析了两个FIP晶体结构(PDB id: 3F3H和3KCW)以及来自不同分类类群的同源序列。我们的研究结果揭示了真菌、细菌和人类序列中的保守基序,表明潜在的功能相似性。比较结构分析强调了FIP结构的显著保存,其变化主要在n端区域。值得注意的是,与细菌毒素的结构一致性,如艰难梭菌的adp核糖基化二元毒素或炭疽芽孢杆菌的炭疽毒素保护性抗原,表明FIP免疫调节作用的机制。gFIPs与免疫相关蛋白(如细菌毒素结合结构域、抗体片段、t细胞受体成分和免疫检查点调节因子(PD-1))之间的结构相似性表明它们可能参与TCR信号通路。这项综合分析阐明了gFIPs多种生物活性的结构基础,并强调了它们作为免疫相关疾病治疗剂的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of structural biology
生物-生化与分子生物学
CiteScore
6.30
自引率
3.30%
发文量
88
审稿时长
65 days
期刊介绍:
Journal of Structural Biology (JSB) has an open access mirror journal, the Journal of Structural Biology: X (JSBX), sharing the same aims and scope, editorial team, submission system and rigorous peer review. Since both journals share the same editorial system, you may submit your manuscript via either journal homepage. You will be prompted during submission (and revision) to choose in which to publish your article. The editors and reviewers are not aware of the choice you made until the article has been published online. JSB and JSBX publish papers dealing with the structural analysis of living material at every level of organization by all methods that lead to an understanding of biological function in terms of molecular and supermolecular structure.
Techniques covered include:
• Light microscopy including confocal microscopy
• All types of electron microscopy
• X-ray diffraction
• Nuclear magnetic resonance
• Scanning force microscopy, scanning probe microscopy, and tunneling microscopy
• Digital image processing
• Computational insights into structure
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


