The Structure of the Apolipoprotein A-I Monomer Provides Insights Into Its Oligomerisation and Lipid-binding Mechanisms
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
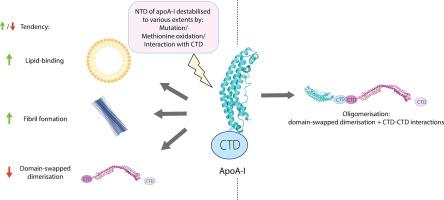
Apolipoprotein A-I (apoA-I) plays important roles in clearing cholesterol and phospholipids from peripheral tissues, forming high-density lipoprotein (HDL). However, despite this important function, apoA-I has a propensity to form amyloid fibrils implicated in atherosclerosis and hereditary amyloidosis. Historically, structural determination of lipid-free or lipid-poor apoA-I has been difficult. Here, we obtained the crystal structure of the apoA-I monomer in complex with the antigen-binding fragment (Fab) of a monoclonal antibody. The structure reveals that the N-terminal domain (NTD, residues 1–184) of apoA-I is a compact four-helical bundle, whereas the C-terminal domain (CTD, residues 185–243) is unresolved in the structure. Molecular Dynamics (MD) simulations and small-angle X-ray scattering (SAXS) analysis revealed that the apoA-I NTD dimerises by domain-swapping and the dimer is elongated. Methionine (Met) oxidation in apoA-I destabilises both full-length apoA-I (apoA-IFL) and C-terminally truncated apoA-I (apoA-IΔ185–243), causing dissociation of the domain-swapped dimer and fibril formation. Met oxidation also increased the lipid-binding ability of apoA-IΔ185–243, while the amyloidogenic mutation, G26R, did not. Hydrogen-deuterium exchange coupled with nuclear magnetic resonance (HDX-NMR), SAXS, and MD analyses showed that triply Met-oxidised (3MetO) and G26R apoA-IΔ185–243 are both highly dynamic but remain partially folded. Based on these results, we propose that domain-swapping dimerisation also exists in apoA-IFL, with the CTD mediating further oligomerisation. We also propose that lipid-binding is promoted by increased global destabilisation in the protein structure, and/or driven by a specific local conformation that is induced by Met-oxidation but not the G26R mutation.
载脂蛋白A-I单体的结构提供了对其寡聚化和脂质结合机制的见解。
载脂蛋白A-I (apoA-I)在清除外周组织中的胆固醇和磷脂,形成高密度脂蛋白(HDL)中起重要作用。然而,尽管有这种重要的功能,apoa - 1有形成淀粉样蛋白原纤维的倾向,与动脉粥样硬化和遗传性淀粉样变性有关。从历史上看,无脂或无脂apoA-I的结构测定一直很困难。在这里,我们获得了apoA-I单体与单克隆抗体抗原结合片段(Fab)配合物的晶体结构。该结构揭示了apoA-I的n端结构域(NTD,残基1-184)是一个紧凑的四螺旋束,而c端结构域(CTD,残基185-243)在该结构中是未解析的。分子动力学(MD)模拟和小角x射线散射(SAXS)分析表明,apoA-I NTD通过结构域交换发生二聚体,二聚体被延长。蛋氨酸(Met)在apoA-I中的氧化使全长apoA-I (apoA-IFL)和c端截断的apoA-I (apoA-IΔ185-243)不稳定,导致结构域交换二聚体的解离和纤维的形成。Met氧化也增加了apoA-IΔ185-243的脂质结合能力,而淀粉样变性突变G26R则没有。氢-氘交换耦合核磁共振(HDX-NMR), SAXS和MD分析表明,三重met氧化(3MetO)和G26R apoA-IΔ185-243都是高度动态的,但仍保持部分折叠。基于这些结果,我们提出在apoA-IFL中也存在结构域交换二聚化,CTD介导进一步的寡聚化。我们还提出,脂质结合是由蛋白质结构的全局不稳定性增加和/或由met氧化诱导的特定局部构象驱动的,而不是由G26R突变引起的。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


