Identification and deciphering novel compounds dynamics against DTYMK: A potential oncogene against pancreatic cancer
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
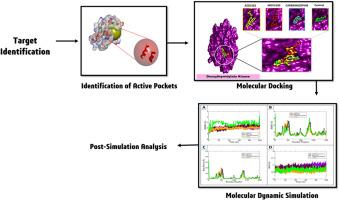
Deoxythymidylate kinase (DTYMK) plays a key role in the progression of pancreatic cancer (PC). The computational study began by selecting DTYMK as an anti-cancer target and using a drug library called the FDA-approved anticancer drug library, which contains 1745 compounds. Using molecular docking analysis, MD simulation, and pharmacokinetics profiling, the study identified three novel drug compounds, AZD2281, MDV3100, and carbamazepine with docking scores of −9.8 kcal/mol, −9.7 kcal/mol, and −9.6 kcal/mol, respectively. Additionally, the control compound (E)-1-(4-(hydroxy (phosphonooxy) phosphoryl) but-2-en-1-yl)-5-methyl pyrimidine-1, 3-diium-2, 4-bis (olate) showed a binding affinity of −7.3 kcal/mol. All three compounds met the Lipinski Rule of Five and were considered drug-like. The DTYMK complexes had mean RMSD values of 2.00 Å, 2.07 Å, 1.92 Å, and 2.84 Å for AZD2281, MDV3100, carbamazepine, and the control, respectively. The AZD2281 complex demonstrated the highest stability with no major deviation observed. Salt bridge analysis revealed key interactions such as Glu140-Arg14, Glu26-Arg22, Glu40-Arg37, and Asp103-Lys106. PCA analysis was performed to simplify large datasets, revealing that the eigenvalue patterns of the complexes were quite distinct. MMGB/PBSA calculations showed values of −95.96 kcal/mol and −97.36 kcal/mol for AZD2281, indicating its stability during ligand binding. The most suitable entropy value for AZD2281 suggested the most consistent and ordered binding. This in silico approach identified novel therapeutic compounds for pancreatic cancer, which still require experimental validation and may be modified for targeting DTYMK.
鉴定和破译新的化合物动力学对抗DTYMK:胰腺癌的潜在致癌基因
脱氧胸腺苷酸激酶(DTYMK)在胰腺癌(PC)的进展中起着关键作用。计算研究首先选择DTYMK作为抗癌靶点,并使用一个被称为fda批准的抗癌药物库的药物库,其中包含1745种化合物。通过分子对接分析、MD模拟和药代动力学分析,该研究确定了三种新型药物化合物AZD2281、MDV3100和卡马西平的对接评分分别为−9.8 kcal/mol、−9.7 kcal/mol和−9.6 kcal/mol。另外,对照化合物(E)-1-(4-(羟基(磷酸)磷基)- 2-烯-1-基)-5-甲基嘧啶- 1,3 -二- 2,4 -双(酸盐)的结合亲和力为−7.3 kcal/mol。这三种化合物都符合利平斯基五法则,被认为是类药物。DTYMK复合物对AZD2281、MDV3100、卡马西平和对照的平均RMSD值分别为2.00 Å、2.07 Å、1.92 Å和2.84 Å。AZD2281配合物表现出最高的稳定性,没有观察到大的偏差。盐桥分析揭示了Glu140-Arg14、Glu26-Arg22、Glu40-Arg37和Asp103-Lys106等关键相互作用。对大型数据集进行主成分分析,发现复合体的特征值模式非常明显。MMGB/PBSA计算显示AZD2281的值分别为- 95.96 kcal/mol和- 97.36 kcal/mol,表明AZD2281在配体结合过程中具有稳定性。最适合AZD2281的熵值表明其结合最一致且有序。这种计算机方法确定了新的胰腺癌治疗化合物,这些化合物仍需要实验验证,并且可能针对DTYMK进行修饰。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


