Structural Basis of Novel Bile Acid-Based Modulators of FXR
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
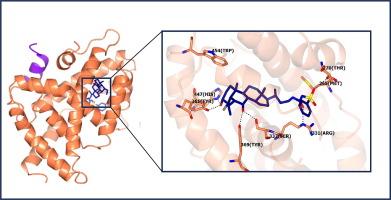
Following its deorphanisation in the early 2000s, the farnesoid X receptor (FXR) attracted significant attention for regulating genes involved in bile acid, lipid and glucose metabolism and inflammation, pathways central to many liver diseases. As such, pharmaceutical efforts targeted FXR for their treatment. However, while FXR agonists, such as obeticholic acid, have been studied in clinical trials, many were associated with adverse effects arising from the promiscuity of systemic FXR activation, thus efforts to limit or selectively modulate the downstream effects of FXR are crucially important. In work here, two novel bile acid derivatives, previously identified via molecular docking and cell-based screening, were validated by X-ray crystallography and tested in LanthaScreen coactivator recruitment assays. Their effects on downstream FXR signalling were assessed in vitro in hepatocellular carcinoma cells, and in vivo in C57BL/6 mice, by RNA sequencing and RT-qPCR. The novel compounds exhibited potent and selective FXR agonist activity. Co-crystal structures of FXR LBD with both compounds, demonstrated distinctive binding modes for each, including occupancy of a receptor sub-pocket associated with allosteric activation, not observed with classic bile acids. Both compounds were up to four-fold more potent than obeticholic acid and demonstrated ligand-dependent differences in coactivator recruitment assays. In vitro, both compounds induced greater changes in the expression of FXR target genes, at lower doses than obeticholic acid. In vivo, compound-dependent differential gene expression was observed. These findings suggest that the novel compounds may enable gene-specific FXR regulation through differential coactivator usage and hold potential to overcome the shortcomings of current bile acid drugs, thus representing promising candidates for further research.
新型胆汁酸基FXR调节剂的结构基础。
在21世纪初其去孤儿化之后,法内酯X受体(FXR)因调节胆汁酸、脂质和葡萄糖代谢和炎症相关基因而引起了极大的关注,这些基因是许多肝脏疾病的核心途径。因此,制药公司将FXR作为他们的治疗目标。然而,虽然FXR激动剂,如欧贝胆酸,已经在临床试验中进行了研究,但许多激动剂与系统FXR激活的混杂引起的不良反应有关,因此限制或选择性调节FXR的下游作用至关重要。在这里的工作中,两种新的胆汁酸衍生物,先前通过分子对接和基于细胞的筛选鉴定,通过x射线晶体学验证,并在LanthaScreen共激活剂招募试验中进行了测试。通过RNA测序和RT-qPCR,在体外肝癌细胞和体内C57BL/6小鼠中评估了它们对下游FXR信号传导的影响。新化合物表现出有效的选择性FXR激动剂活性。FXR LBD与这两种化合物的共晶结构显示出每种化合物的独特结合模式,包括占用与变构活化相关的受体子口袋,这在经典胆汁酸中没有观察到。这两种化合物的效力都比奥贝胆酸高4倍,并且在共激活剂招募试验中表现出配体依赖性差异。在体外实验中,这两种化合物在较低剂量下诱导FXR靶基因表达的更大变化。在体内,观察到化合物依赖性差异基因表达。这些发现表明,这些新化合物可能通过不同的辅激活剂使用来实现基因特异性FXR调控,并具有克服当前胆汁酸药物缺点的潜力,因此具有进一步研究的前景。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


