Generative AI techniques for conformational diversity and evolutionary adaptation of proteins
IF 6.1
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The advent of AlphaFold and consumer large language models have elicited unprecedented development of artificial intelligence (AI). AI has had substantial impact in every area of research, including in molecular biology. This is principally in thanks to contributions to the Protein Data Bank and various genome sequence databases, providing an astronomical amount of data for model training. These databases contain evolutionary information explicitly and implicitly, allowing accurate predictions and deep insights into biological questions. Here, we describe recent state-of-the-art applications of AI that exploit evolutionary relationships. This includes structure prediction and design, conformational ensemble generation, and functional site identification. We present a brief snapshot of AI usage in studying protein structure and dynamics, a field that is advancing at breakneck speed.
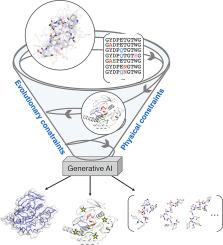
蛋白质构象多样性和进化适应的生成人工智能技术
AlphaFold和消费级大语言模型的出现,引发了人工智能(AI)的空前发展。人工智能在包括分子生物学在内的每个研究领域都产生了重大影响。这主要是由于对蛋白质数据库和各种基因组序列数据库的贡献,为模型训练提供了天文数字的数据量。这些数据库明确地或隐含地包含了进化信息,允许对生物学问题进行准确的预测和深入的了解。在这里,我们描述了利用进化关系的人工智能的最新应用。这包括结构预测和设计、构象集合生成和功能位点识别。我们简要介绍了人工智能在研究蛋白质结构和动力学方面的应用,这一领域正在以惊人的速度发展。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current opinion in structural biology
生物-生化与分子生物学
CiteScore
12.20
自引率
2.90%
发文量
179
审稿时长
6-12 weeks
期刊介绍:
Current Opinion in Structural Biology (COSB) aims to stimulate scientifically grounded, interdisciplinary, multi-scale debate and exchange of ideas. It contains polished, concise and timely reviews and opinions, with particular emphasis on those articles published in the past two years. In addition to describing recent trends, the authors are encouraged to give their subjective opinion of the topics discussed.
In COSB, we help the reader by providing in a systematic manner:
1. The views of experts on current advances in their field in a clear and readable form.
2. Evaluations of the most interesting papers, annotated by experts, from the great wealth of original publications.
[...]
The subject of Structural Biology is divided into twelve themed sections, each of which is reviewed once a year. Each issue contains two sections, and the amount of space devoted to each section is related to its importance.
-Folding and Binding-
Nucleic acids and their protein complexes-
Macromolecular Machines-
Theory and Simulation-
Sequences and Topology-
New constructs and expression of proteins-
Membranes-
Engineering and Design-
Carbohydrate-protein interactions and glycosylation-
Biophysical and molecular biological methods-
Multi-protein assemblies in signalling-
Catalysis and Regulation
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


