Unraveling the molecular interaction of Larotrectinib with calf thymus DNA: A comprehensive study using multi-spectroscopic, thermodynamic, and computational techniques
IF 2.2
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
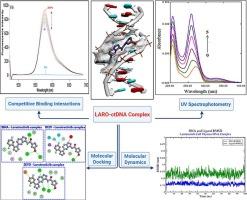
The study of the interaction between small molecules and biological macromolecules is a critical area of research with significant implications across various scientific fields. Larotrectinib, a tropomyosin kinase inhibitor, is used to treat patients with solid tumors harboring neurotrophic tyrosine receptor kinase (NTRK) gene fusions. In this investigation, the interaction between larotrectinib and calf thymus DNA (ctDNA) was thoroughly examined using a combination of techniques, including UV–Vis spectrophotometry, spectrofluorimetry, viscosity measurements, ionic strength variation, thermodynamic analysis, molecular dynamics simulations, and docking studies. The results demonstrated a strong binding interaction between larotrectinib and ctDNA, with the drug primarily binding to the minor groove of ctDNA. This binding mode was established through competitive binding assays using ethidium bromide and rhodamine B, as well as UV–Vis spectroscopy and viscosity analysis. The binding constant (Kb) at 298 K, determined using the Benesi-Hildebrand equation, was found to be 4.4 × 105 M−1, pointing out a high binding affinity between larotrectinib and ctDNA. Thermodynamic analysis revealed that the interaction is driven mainly by hydrophobic forces and hydrogen bonding, as evidenced by the calculated enthalpy (ΔH0) and entropy (ΔS0) changes. Molecular docking studies further supported these findings, showing that larotrectinib binds preferentially to the AT-rich regions of the B-DNA minor groove. This was validated by molecular dynamics studies, which provided additional confirmation of the binding mechanism. Overall, these findings provide valuable understanding into the molecular interactions and pharmacological mechanisms of larotrectinib, contributing to a deeper insight of its role as a potent anticancer agent.
揭示larorectinib与小牛胸腺DNA的分子相互作用:一项使用多光谱、热力学和计算技术的综合研究
小分子与生物大分子之间相互作用的研究是一个重要的研究领域,在各个科学领域都具有重要的意义。larorectinib是一种原肌球蛋白激酶抑制剂,用于治疗含有神经营养酪氨酸受体激酶(NTRK)基因融合的实体瘤患者。在这项研究中,larorectinib和小牛胸腺DNA (ctDNA)之间的相互作用进行了全面的研究,包括紫外可见分光光度法、荧光光谱法、粘度测量、离子强度变化、热力学分析、分子动力学模拟和对接研究。结果表明larorectinib与ctDNA之间具有很强的结合相互作用,药物主要结合在ctDNA的小凹槽上。这种结合模式是通过溴化乙锭和罗丹明B的竞争结合实验,以及紫外可见光谱和粘度分析建立的。利用Benesi-Hildebrand方程确定larorectinib与ctDNA在298 K时的结合常数(Kb)为4.4 × 105 M−1,表明larorectinib与ctDNA具有较高的结合亲和力。热力学分析表明,相互作用主要由疏水力和氢键驱动,计算焓(ΔH0)和熵(ΔS0)的变化证明了这一点。分子对接研究进一步支持了这些发现,表明larorectinib优先结合B-DNA次要凹槽中富含at的区域。分子动力学研究证实了这一点,进一步证实了其结合机制。总的来说,这些发现为larorectinib的分子相互作用和药理机制提供了有价值的理解,有助于更深入地了解其作为一种有效的抗癌药物的作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Biophysical chemistry
生物-生化与分子生物学
CiteScore
6.10
自引率
10.50%
发文量
121
审稿时长
20 days
期刊介绍:
Biophysical Chemistry publishes original work and reviews in the areas of chemistry and physics directly impacting biological phenomena. Quantitative analysis of the properties of biological macromolecules, biologically active molecules, macromolecular assemblies and cell components in terms of kinetics, thermodynamics, spatio-temporal organization, NMR and X-ray structural biology, as well as single-molecule detection represent a major focus of the journal. Theoretical and computational treatments of biomacromolecular systems, macromolecular interactions, regulatory control and systems biology are also of interest to the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


