METACONE: A scalable framework for exploring the conversion cone of metabolic networks
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
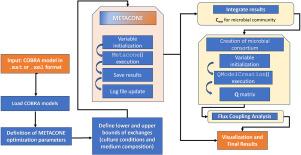
Elementary Conversion Modes (ECMs) – a subset of Elementary Flux Modes (EFMs) – capture the entire production/consumption potential of a metabolic network, providing a more practical view of its interactions with the environment. Despite its reduced size, the set of ECMs is too large for exhaustive enumeration in models reaching genome scale. To address this limitation, we have developed METACONE (METAbolic Conversion cOne for Network Exploration), a scalable algorithm for the computation of a representative linear basis of the conversion cone, the subspace in which all ECMs lie. Two METACONE variants are proposed based on the solution of a series of linear programs following different heuristics. We evaluated the performance of the variants on metabolic models of different sizes, demonstrating their scalability. We further analyzed the resulting basis to explore metabolic capabilities under different environmental conditions in Escherichia coli, identifying metabolic patterns consistent with reported data. Finally, we applied the algorithm to explore metabolic interactions in a microbial consortium of Phocaeicola dorei and Lachnoclostridium symbiosum, recapitulating known cross-feeding interactions and suggesting new possibilities. We envision METACONE as a valuable tool for understanding microbial metabolism in increasingly complex consortia while addressing current scalability challenges.
METACONE:一个可扩展的框架,用于探索代谢网络的转换锥
基本转换模式(ecm)是基本通量模式(efm)的一个子集,它捕获了代谢网络的整个生产/消费潜力,为其与环境的相互作用提供了更实用的观点。尽管ecm的大小减小了,但在达到基因组规模的模型中,ecm的集合太大了,无法进行详尽的枚举。为了解决这一限制,我们开发了METACONE(网络探索代谢转换锥),这是一种可扩展的算法,用于计算转换锥的代表性线性基,即所有ecm所在的子空间。基于不同启发式方法求解一系列线性规划,提出了METACONE的两种变体。我们评估了这些变体在不同大小的代谢模型上的性能,证明了它们的可扩展性。我们进一步分析了结果基础,以探索大肠杆菌在不同环境条件下的代谢能力,确定与报道数据一致的代谢模式。最后,我们应用该算法探索了Phocaeicola dorei和共生Lachnoclostridium symbiosum微生物联合体的代谢相互作用,概述了已知的交叉摄食相互作用,并提出了新的可能性。我们设想METACONE作为一个有价值的工具,在日益复杂的联盟中了解微生物代谢,同时解决当前的可扩展性挑战。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


