Towards methylation-based redefinition of TAL1 positive T-cell acute lymphoblastic leukaemia (T-ALL)
IF 13.4
1区 医学
Q1 HEMATOLOGY
引用次数: 0
Abstract
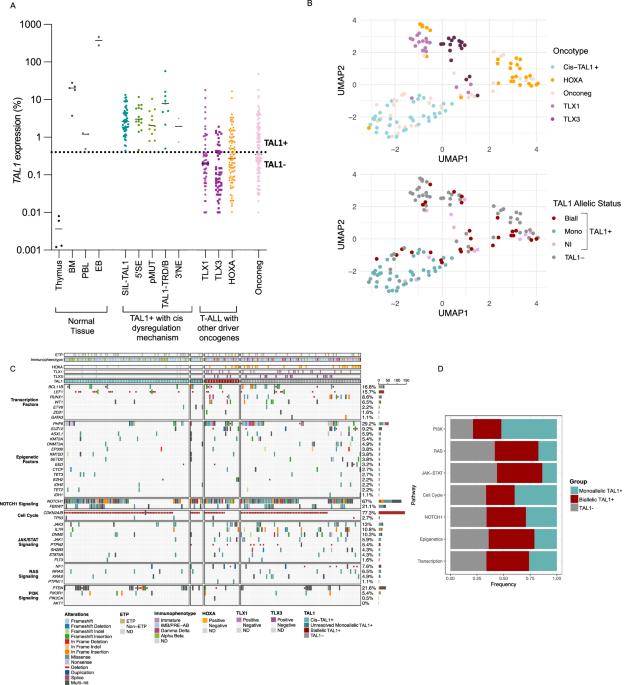
TAL1 is one of the most frequently dysregulated oncogenes in T-cell Acute Lymphoblastic Leukaemia (T-ALL). However, the precise frequency and prognostic impact associated with its dysregulation remains unclear and is confounded by TAL1’s diverse dysregulation mechanisms. TAL1 dysregulation is detected by TAL1 transcript quantification, though this technique may be subject to interference by TAL1 transcripts deriving from residual haematological cells that physiologically express high levels of the gene. We hypothesised TAL1 DNA methylation could provide a more reliable biomarker than TAL1 transcript quantification alone. We extensively studied TAL1 dysregulation in a large adult and paediatric T-ALL cohort (n = 401) and designed a TAL1 specific MS-MLPA assay to determine methylation levels. Whereas monoallelic TAL1 + T-ALL had homogeneous gene expression profiles, never expressed other driver oncogenes and were TAL1 hypomethylated (methylation ratio <0.4), biallelic TAL1 + T-ALL were enriched in expression of other driver oncogenes (TLX1, TLX3, HOXA), and had heterogeneous transcriptomes and TAL1 methylation levels. In PDX analysis, monoallelic TAL1 expression was stable, contrary to biallelic expression which mostly derived from residual non-malignant haematopoietic cells. Importantly, we report 5 novel TAL1 dysregulation mechanisms using long-read nanopore and OGM analysis, and show that TAL1 hypomethylation identifies TAL1 dysregulation, and is associated with worse prognosis.
基于甲基化的TAL1阳性t细胞急性淋巴细胞白血病(T-ALL)的重新定义
TAL1是t细胞急性淋巴细胞白血病(T-ALL)中最常见的失调癌基因之一。然而,与TAL1失调相关的准确频率和预后影响仍不清楚,并与TAL1的多种失调机制相混淆。TAL1的失调是通过TAL1转录物定量检测的,尽管这项技术可能会受到来自残余血液学细胞的TAL1转录物的干扰,这些细胞在生理上表达高水平的TAL1基因。我们假设TAL1 DNA甲基化可以提供比单独TAL1转录物定量更可靠的生物标志物。我们在一个大型成人和儿童T-ALL队列(n = 401)中广泛研究了TAL1的失调,并设计了一种TAL1特异性的MS-MLPA检测方法来确定甲基化水平。单等位基因TAL1 + T-ALL具有均匀的基因表达谱,从不表达其他驱动癌基因,TAL1低甲基化(甲基化率<;0.4),而双等位基因TAL1 + T-ALL则富含其他驱动癌基因(TLX1, TLX3, HOXA)的表达,并且具有异质性的转录组和TAL1甲基化水平。在PDX分析中,单等位基因的TAL1表达是稳定的,而双等位基因的TAL1表达主要来源于残留的非恶性造血细胞。重要的是,我们使用长读纳米孔和OGM分析报告了5种新的TAL1失调机制,并表明TAL1低甲基化识别TAL1失调,并与较差的预后相关。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


