Comparative Study of a Variant Neural Relational Inference Deep Learning Model and Dynamical Network Analysis for p53-DNA Allosteric Interactions
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
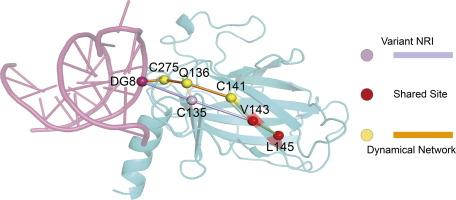
Protein allostery is a critical regulatory mechanism in various biological processes, representing a challenging aspect of biological research. Dynamical network analysis serves as a foundational computational methodology for studying allosteric effects, while the recent emergence of neural relational inference (NRI) model has introduced novel insights into understanding it. In this study, we modified NRI model by integrating the multi-head self-attention module of transformer, and then compared the variant model with dynamical network analysis and initial NRI model for p53-DNA allosteric interactions. Our results show that the variant model is more focused on long-range allosteric than dynamical network in predicting p53-DNA allosteric pathways, and enhances superior accuracy and comprehensiveness compared to initial NRI model. Moreover, divergent allosteric pathways in wild-type (WT) versus mutant (MT) p53 may underlie their substantially different DNA recognition and binding behaviors. Finally, we found that even with undefined ending nodes, allosteric pathways consistently propagate from mutation sites toward DNA, and the length of pathways in MT p53 is significantly longer than that of WT p53. These suggest that mutation sites impair long-range allosteric communication, potentially disrupting signal transmission efficiency. Our results offer novel insights for a deep understanding of protein allosteric pathways through different methods.
变异神经关系推理深度学习模型与p53-DNA变构相互作用动态网络分析的比较研究。
蛋白质变构是多种生物过程中的重要调控机制,是生物学研究的一个具有挑战性的方面。动态网络分析是研究变构效应的基础计算方法,而最近出现的神经关系推理(NRI)模型为理解变构效应提供了新的见解。本研究通过整合变压器多头自注意模块对NRI模型进行改进,并将该模型与动态网络分析和初始NRI模型进行了比较,分析了p53-DNA变构相互作用。结果表明,该变异模型在预测p53-DNA变构途径时更侧重于远程变构而非动态网络,与初始NRI模型相比,具有更高的准确性和全面性。此外,野生型(WT)和突变型(MT) p53的不同变构途径可能是它们本质上不同的DNA识别和结合行为的基础。最后,我们发现,即使没有明确的结束节点,变构通路也始终从突变位点向DNA传播,MT p53的通路长度明显长于WT p53。这表明突变位点损害了远程变构通信,潜在地破坏了信号传输效率。我们的结果为通过不同的方法深入了解蛋白质变构途径提供了新的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


