Harnessing transcriptome data of Viola inconspicua for discovery of novel cyclotides and AEP ligases
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
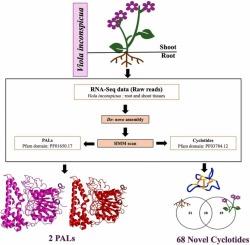
Cyclotides are plant-derived cyclic peptides with three conserved disulfide linkages, forming a cyclic cystine knot (CCK) motif. This CCK topology makes them ultra stable structures and resistant to thermal and chemical degradation. Cyclotides are known to exhibit, anti-HIV, antimicrobial, cytotoxic, hemolytic and pesticidal bioactivities. They have been reported in six angiosperm families (Cucurbitaceae, Fabaceae, Poaceae, Rubiaceae, Solanaceae, Violaceae). The identification of novel cyclotides is the first important step for investigating their potential applications in agriculture and therapeutics. To address this need, the present study employed a de novo transcriptome assembly of Viola inconspicua root and shoot tissues. HMM-based searches were used to identify novel cyclotides and the enzymes involved in their biosynthesis, specifically asparaginyl endopeptidases (AEPs). The analysis revealed six types of cyclotide precursor (CP) gene architectures and 68 novel cyclotides of which 31 and 19 were exclusive to roots and shoots respectively, and 18 were common in both tissues. Assessment of potential bioactivity of 68 novel cyclotides was investigated by analysing their physicochemical characteristics, loop sequence variations and phylogenetic studies. Furthermore, the analysis revealed 40 AEP isoforms. Two of these were identified as potential peptide asparaginyl ligases (PALs), important for the cyclization of cyclotides. Moreover, comparison of homology models of potential PALs with experimentally validated structure of PAL from Viola yedoensis (VyPAL2) revealed high structural homology. In summary, this study reveals tissue specific diversity of cyclotides in V. inconspicua; identifies novel cyclotides, provides insights on CP gene architectures and structure of potential PALs.
利用紫堇转录组数据发现新的环核苷酸和AEP连接酶
环肽是植物衍生的环状肽,具有三个保守的二硫键,形成环胱氨酸结(CCK)基序。这种CCK拓扑结构使它们具有超稳定的结构,并且耐热和化学降解。已知环聚糖具有抗艾滋病毒、抗菌、细胞毒性、溶血和杀虫的生物活性。在被子植物科(葫芦科、豆科、豆科、茜草科、茄科、堇菜科)中已有报道。鉴定新的环核苷酸是研究其在农业和治疗学中的潜在应用的重要的第一步。为了解决这一需求,本研究采用了紫堇根和茎组织的从头转录组组装。基于hmm的搜索用于鉴定新的环核苷酸和参与其生物合成的酶,特别是天冬酰胺内肽酶(AEPs)。共发现6种环肽前体(CP)基因结构,68种新环肽,其中31种和19种分别为根和芽所特有,18种为根和芽所共有。通过分析68种新型环核苷酸的理化特性、环序列变异和系统发育研究,对其潜在生物活性进行了评价。此外,分析还发现了40个AEP亚型。其中两个被鉴定为潜在的肽天冬酰胺酰连接酶(PALs),对环核苷酸的环化很重要。此外,潜在PAL的同源性模型与实验验证的紫堇(Viola yedoensis) PAL (VyPAL2)的结构进行了比较,显示出高度的结构同源性。综上所述,本研究揭示了环核苷酸的组织特异性多样性;鉴定新的环核苷酸,提供对CP基因结构和潜在pal结构的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


