The complete mitochondrial genome assembly of Bali cattle (Bos javanicus) using Oxford nanopore long-read sequencing technology
IF 2.4
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
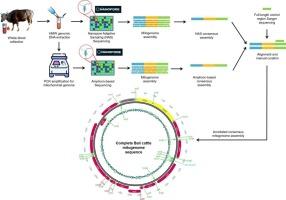
Bali cattle, domesticated from wild banteng (Bos javanicus), serve as an important indigenous genetic resource in Indonesia. Creating a detailed mitochondrial genome (mitogenome) sequence of Bali cattle is essential for population genetic research and promoting sustainable breeding. This study aimed to assemble the complete mitogenome sequence of Bali cattle, analyze its genetic features, and explore its phylogenetic position within the Bos genus. High-molecular-weight genomic DNA was extracted from the blood of a male Bali cattle, and mitogenome sequencing was performed using Oxford Nanopore long-read sequencing, combining nanopore adaptive sampling (NAS) and amplicon-based sequencing approaches. Both NAS and amplicon-based methods can assemble a complete mitogenome of Bali cattle. However, validating NAS assemblies through amplicon-based sequencing improves accuracy and completeness. The complete mitogenome of Bali cattle spans 16,387 bp, comprising 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a non-coding control region (CR), which showed a structure similar to that observed in other Bos species. Phylogenetic analysis showed that Bali cattle group with B. javanicus, closely related to Javan banteng (B. j. javanicus). Interestingly, a unique tandem repeat sequence was found only in the CR of B. j. javanicus, including Bali cattle. Notably, the current NCBI reference mitogenome for B. javanicus (NC_012706.1) is more closely related to B. taurus than to verified B. javanicus sequences, underscoring the need for a reliable B. javanicus-specific reference mitogenome. This study offers novel insights into the comprehensive mitogenomic architecture of Bali cattle. Furthermore, the discovery of a unique tandem repeat motif offers potential as a marker for Bali cattle identification and its applications in forensic and product authentication.
利用牛津纳米孔长读测序技术对巴厘牛(Bos javanicus)线粒体基因组进行完整组装
巴厘岛牛是从野生班腾(Bos javanicus)驯化而来,是印度尼西亚重要的土著遗传资源。建立巴厘牛线粒体基因组(有丝分裂基因组)的详细序列对于种群遗传研究和促进可持续育种至关重要。本研究旨在收集巴厘牛有丝分裂全基因组序列,分析其遗传特征,并探讨其在牛属中的系统发育位置。从一头雄性巴厘牛的血液中提取高分子量基因组DNA,结合纳米孔自适应采样(NAS)和基于扩增子的测序方法,采用牛津纳米孔长读测序技术进行有丝分裂基因组测序。NAS和基于扩增子的方法均可组装巴厘牛的完整有丝分裂基因组。然而,通过基于扩增子的测序来验证NAS组装可以提高准确性和完整性。巴厘牛有丝分裂基因组全长16387 bp,包括13个蛋白质编码基因(PCGs)、22个转移RNA (tRNA)基因、2个核糖体RNA (rRNA)基因和1个非编码控制区(CR),结构与其他牛属相似。系统发育分析表明,巴厘牛群具有爪哇白牛,与爪哇班腾(Javan banteng)亲缘关系较近。有趣的是,一个独特的串联重复序列仅在包括巴厘牛在内的爪哇牛的CR中被发现。值得注意的是,目前的NCBI参考爪哇芽孢杆菌有丝分裂基因组(NC_012706.1)与金牛芽孢杆菌的关系比已验证的爪哇芽孢杆菌序列更密切,这表明需要一个可靠的爪哇芽孢杆菌特异性参考有丝分裂基因组。这项研究为巴厘牛的全面有丝分裂基因组结构提供了新的见解。此外,独特的串联重复基序的发现为巴厘牛鉴定及其在法医和产品鉴定中的应用提供了潜在的标记。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Gene
生物-遗传学
CiteScore
6.10
自引率
2.90%
发文量
718
审稿时长
42 days
期刊介绍:
Gene publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


